Developing an Integrated Remote Sensing Based Biodiversity Index for Predicting Animal Species Richness
Abstract
:1. Introduction
- (1)
- What are the differences between the distributions of various animal classes (mammals, birds, and amphibians) on a global scale?
- (2)
- What are the correlations between remote sensing metrics and species richness distributions on a global scale?
- (3)
- Given the major driving metrics, can we develop a multivariate integration index to map the global species richness continuously?
2. Data and Methods
2.1. Data
2.1.1. Species Richness Data
2.1.2. Remote Sensing Data
2.1.3. Biome Data
2.2. Methods
2.2.1. Data Integration and Standardization
2.2.2. Regression Analysis
2.2.3. The Akaike Information Criterion
2.2.4. Three Machine Learning Methods for Metrics Integration
3. Results
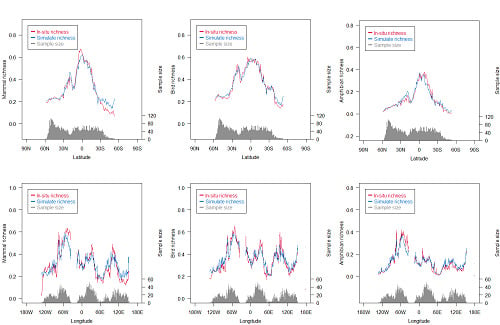
3.1. Richness Distribution of Three Animal Classes
3.2. Characterization of Various Remote Sensing Metrics
3.3. Integration of Various Remote Sensing Metrics
4. Discussion
4.1. Differences in Distribution of the Three Animal Classes
4.2. Attribution of Differences between Remote Sensing Metrics
4.3. Integration of Various Remote Sensing Metrics
4.4. Limitation and Recommendation for Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.; Jongman, R.; Scholes, R.; Bruford, M.W.; Brummitt, N.; Butchart, S.; Cardoso, A. Essential biodiversity variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimm, S.L.; Jenkins, C.N.; Abell, R.G.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef] [PubMed]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Newbold, T.; Hudson, L.N.; Hill, S.L.L.; Contu, S.; Lysenko, I.; Senior, R.A.; Boerger, L.; Bennett, D.J.; Choimes, A.; Collen, B.; et al. Global effects of land use on local terrestrial biodiversity. Nature 2015, 520, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Butchart, S.H.; Walpole, M.; Collen, B.; Van Strien, A.; Scharlemann, J.P.; Almond, R.E.; Baillie, J.E.; Bomhard, B.; Brown, C.; Bruno, J. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- Noss, R.F. Indicators for monitoring biodiversity: A hierarchical approach. Conserv. Biol. 1990, 4, 355–364. [Google Scholar] [CrossRef]
- Purvis, A.; Hector, A. Getting the measure of biodiversity. Nature 2000, 405, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Jetz, W.; Rahbek, C. Geographic range size and determinants of avian species richness. Science 2002, 297, 1548–1551. [Google Scholar] [CrossRef] [PubMed]
- Kreft, H.; Jetz, W. Global patterns and determinants of vascular plant diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 5925–5930. [Google Scholar] [CrossRef] [PubMed]
- Guenard, B.; Weiser, M.D.; Dunn, R.R. Global models of ant diversity suggest regions where new discoveries are most likely are under disproportionate deforestation threat. Proc. Natl. Acad. Sci. USA 2012, 109, 7368–7373. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.N.; Guenard, B.; Diamond, S.E.; Weiser, M.D.; Dunn, R.R. Conservation implications of divergent global patterns of ant and vertebrate diversity. Divers. Distrib. 2013, 19, 1084–1092. [Google Scholar] [CrossRef]
- Costanza, R.; Fisher, B.; Mulder, K.; Liu, S.; Christopher, T. Biodiversity and ecosystem services: A multi-scale empirical study of the relationship between species richness and net primary production. Ecol. Econ. 2007, 61, 478–491. [Google Scholar] [CrossRef]
- Hurlbert, A.H.; Jetz, W. Species richness, hotspots, and the scale dependence of range maps in ecology and conservation. Proc. Natl. Acad. Sci. USA 2007, 104, 13384–13389. [Google Scholar] [CrossRef] [PubMed]
- Pardieck, K.L.; Ziolkowski, D.J., Jr.; Lutmerding, M.; Campbell, K.; Hudson, M.-A.R. North American Breeding Bird Survey Dataset 1966-2013, Version 2016.0; US Geological Survey: Reston, VA, USA; Patuxent Wildlife Research Center: Laurel, MD, USA, 2017; Available online: www.pwrc.usgs.gov/BBS/RawData/ (accessed on 8 May 2018).
- Constable, H.; Guralnick, R.; Wieczorek, J.; Spencer, C.L.; Peterson, A.T. Vertnet: A new model for biodiversity data sharing. PLoS Biol. 2010, 8, e1000309. [Google Scholar] [CrossRef] [PubMed]
- Garciarosello, E.; Guisande, C.; Heine, J.; Pelayovillamil, P.; Manjarreshernandez, A.; Vilas, L.G.; Gonzalezdacosta, J.; Vaamonde, A.; Granadolorencio, C. Using modestr to download, import and clean species distribution records. Methods Ecol. Evol. 2014, 5, 708–713. [Google Scholar] [CrossRef]
- IUCN. The Iucn Red List of Threatened Species. 2017. Available online: http://www.iucnredlist.org/ (accessed on 8 May 2018).
- GBIF. Gbif Data Portal. 2017. Available online: http://www.gbif.org/ (accessed on 8 May 2018).
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2001, 22, 2377–2400. [Google Scholar] [CrossRef]
- Gougeon, F.A. Comparison of possible multispectral classification schemes for tree crowns individually delineatedon high spatial resolution meis images. Can. J. Remote Sens. 1995, 21, 1–9. [Google Scholar] [CrossRef]
- Kier, G.; Kreft, H.; Lee, T.M.; Jetz, W.; Ibisch, P.L.; Nowicki, C.; Mutke, J.; Barthlott, W. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. USA 2009, 106, 9322–9327. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, C.; Graves, G.R. Multiscale assessment of patterns of avian species richness. Proc. Natl. Acad. Sci. USA 2001, 98, 4534–4539. [Google Scholar] [CrossRef] [PubMed]
- Tuanmu, M.N.; Jetz, W. A global, remote sensing-based characterization of terrestrial habitat heterogeneity for biodiversity and ecosystem modelling. Glob. Ecol. Biogeogr. 2015, 24, 1329–1339. [Google Scholar] [CrossRef]
- Duro, D.C.; Coops, N.C.; Wulder, M.A.; Han, T. Development of a large area biodiversity monitoring system driven by remote sensing. Prog. Phys. Geogr. 2007, 31, 235–260. [Google Scholar] [CrossRef]
- Currie, D.J. Energy and large-scale patterns of animal- and plant-species richness. Am. Nat. 1991, 137, 27–49. [Google Scholar] [CrossRef]
- Rosenzweig, M.L.; Abramsky, Z. Species Diversity in Ecological Communities; University of Chicago Press: Chicago, IL, USA, 1993. [Google Scholar]
- Liang, J.; Crowther, T.W.; Picard, N.; Wiser, S.; Zhou, M.; Alberti, G.; Schulze, E.-D.; McGuire, A.D.; Bozzato, F.; Pretzsch, H.; et al. Positive biodiversity-productivity relationship predominant in global forests. Science 2016, 354, aaf8957. [Google Scholar] [CrossRef] [PubMed]
- Nagalingum, N.S.; Knerr, N.; Laffan, S.W.; González-Orozco, C.E.; Thornhill, A.H.; Miller, J.T.; Mishler, B.D. Continental scale patterns and predictors of fern richness and phylogenetic diversity. Front. Genet. 2015, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.M.; Jetz, W. Remotely sensed high-resolution global cloud dynamics for predicting ecosystem and biodiversity distributions. PLoS Biol. 2016, 14, e1002415. [Google Scholar] [CrossRef] [PubMed]
- Currie, D.J.; Mittelbach, G.G.; Cornell, H.V.; Field, R.; Guégan, J.F.; Hawkins, B.A.; Kaufman, D.M.; Kerr, J.T.; Oberdorff, T.; O’Brien, E. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol. Lett. 2004, 7, 1121–1134. [Google Scholar] [CrossRef]
- Hansson, L.; Fahrig, L.; Merriam, G. Mosaic Landscapes and Ecological Processes; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Guegan, J.-F.; Lek, S.; Oberdorff, T. Energy availability and habitat heterogeneity predict global riverine fish diversity. Nature 1998, 391, 382–384. [Google Scholar] [CrossRef]
- Wright, D.H. Species-energy theory: An extension of species-area theory. Oikos 1983, 41, 496–506. [Google Scholar] [CrossRef]
- Brown, J.H. Two decades of homage to santa rosalia: Toward a general theory of diversity. Integr. Comp. Biol. 1981, 21, 877–888. [Google Scholar] [CrossRef]
- Hutchinson, G.E. Homage to santa rosalia or why are there so many kinds of animals. Am. Nat. 1959, 93, 145–159. [Google Scholar] [CrossRef]
- Hobi, M.L.; Dubinin, M.; Graham, C.H.; Coops, N.C.; Clayton, M.K.; Pidgeon, A.M.; Radeloff, V.C. A comparison of dynamic habitat indices derived from different modis products as predictors of avian species richness. Remote Sens. Environ. 2017, 195, 142–152. [Google Scholar] [CrossRef]
- Menendez, R.; Megias, A.G.; Hill, J.K.; Braschler, B.; Willis, S.G.; Collingham, Y.C.; Fox, R.; Roy, D.B.; Thomas, C.D. Species richness changes lag behind climate change. Proc. R. Soc. B Biol. Sci. 2006, 273, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, N.; Bohninggaese, K. Potential impact of global climate change on species richness of long-distance migrants. Conserv. Biol. 2003, 17, 577–586. [Google Scholar] [CrossRef]
- Lawton, J.H. Plant architecture and the diversity of phytophagous insects. Annu. Rev. Entomol. 1983, 28, 23–39. [Google Scholar] [CrossRef]
- McCoy, E.D.; Bell, S.S. Habitat structure: The evolution and diversification of a complex topic. In Habitat Structure; Springer: Berlin, Germany, 1991; pp. 3–27. [Google Scholar]
- Tews, J.; Brose, U.; Grimm, V.; Tielbörger, K.; Wichmann, M.; Schwager, M.; Jeltsch, F. Animal species diversity driven by habitat heterogeneity/diversity: The importance of keystone structures. J. Biogeogr. 2004, 31, 79–92. [Google Scholar] [CrossRef]
- Skidmore, A.K.; Pettorelli, N. Agree on biodiversity metrics to track from space. Nature 2015, 523, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Scholes, R.; Biggs, R. A biodiversity intactness index. Nature 2005, 434, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Petrou, Z.I.; Manakos, I.; Stathaki, T. Remote sensing for biodiversity monitoring: A review of methods for biodiversity indicator extraction and assessment of progress towards international targets. Biodivers. Conserv. 2015, 24, 2333–2363. [Google Scholar] [CrossRef]
- Buchanan, G.M.; Nelson, A.; Mayaux, P.; Hartley, A.; Donald, P.F. Delivering a global, terrestrial, biodiversity observation system through remote sensing. Conserv. Biol. 2009, 23, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Luoto, M. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inform. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Jenkins, C.N.; Pimm, S.L.; Joppa, L.N. Global patterns of terrestrial vertebrate diversity and conservation. Proc. Natl. Acad. Sci. USA 2013, 110, E2602–E2610. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.N.; Van Houtan, K.S.; Pimm, S.L.; Sexton, J.O. Us protected lands mismatch biodiversity priorities. Proc. Natl. Acad. Sci. USA 2015, 112, 5081–5086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cai, D.; Guo, S.; Guan, Y.; Fraedrich, K.; Nie, Y.; Liu, X.; Bian, X. Spatial-temporal dynamics of china’s terrestrial biodiversity: A dynamic habitat index diagnostic. Remote Sens. 2016, 8, 227. [Google Scholar] [CrossRef]
- Golestani, A.; Gras, R. Using Machine Learning Techniques for Identifying Important Characteristics to Predict Changes in Species Richness in Ecosim, an Individual-Based Ecosystem Simulation. In Proceedings of the World Congress on Engineering and Computer Science, San Francisco, CA, USA, 24–26 October 2012. [Google Scholar]
- Olaya-Marín, E.J.; Martínez-Capel, F.; Vezza, P. A comparison of artificial neural networks and random forests to predict native fish species richness in mediterranean rivers. Knowl. Manag. Aquat. Ecosyst. 2013. [Google Scholar] [CrossRef]
- Golestani, A.; Gras, R. Regularity analysis of an individual-based ecosystem simulation. Chaos 2010, 20, 043120. [Google Scholar] [CrossRef] [PubMed]
- Devaurs, D.; Gras, R. Species abundance patterns in an ecosystem simulation studied through fisher’s logseries. Simul. Model. Pract. Theory 2010, 18, 100–123. [Google Scholar] [CrossRef] [Green Version]
- Mouton, A.M.; Alcaraz-Hernández, J.D.; De Baets, B.; Goethals, P.L.; Martínez-Capel, F. Data-driven fuzzy habitat suitability models for brown trout in spanish mediterranean rivers. Environ. Model. Softw. 2011, 26, 615–622. [Google Scholar] [CrossRef]
- Leclere, J.; Oberdorff, T.; Belliard, J.; Leprieur, F. A comparison of modeling techniques to predict juvenile 0+ fish species occurrences in a large river system. Ecol. Inform. 2011, 6, 276–285. [Google Scholar] [CrossRef]
- Knudby, A.; LeDrew, E.; Brenning, A. Predictive mapping of reef fish species richness, diversity and biomass in zanzibar using ikonos imagery and machine-learning techniques. Remote Sens. Environ. 2010, 114, 1230–1241. [Google Scholar] [CrossRef]
- IUCN. The Iucn Red List of Threatened Species. Version 2010-4. 2010, Volume 9. Available online: http://www.iucnredlist.org/info/categories_criteria2001.html (accessed on 12 November 2017).
- Zhao, M.; Heinsch, F.A.; Nemani, R.R.; Running, S.W. Improvements of the modis terrestrial gross and net primary production global data set. Remote Sens. Environ. 2005, 95, 164–176. [Google Scholar] [CrossRef]
- Coops, N.C.; Wulder, M.A.; Iwanicka, D. Demonstration of a satellite-based index to monitor habitat at continental-scales. Ecol. Indic. 2009, 9, 948–958. [Google Scholar] [CrossRef]
- Coops, N.C.; Wulder, M.A.; Duro, D.C.; Han, T.; Berry, S. The development of a canadian dynamic habitat index using multi-temporal satellite estimates of canopy light absorbance. Ecol. Indic. 2008, 8, 754–766. [Google Scholar] [CrossRef]
- Perez, L.; Nelson, T.A.; Coops, N.C.; Fontana, F.; Drever, C.R. Characterization of spatial relationships between three remotely sensed indirect indicators of biodiversity and climate: A 21years’ data series review across the canadian boreal forest. Int. J. Digit. Earth 2016, 9, 676–696. [Google Scholar] [CrossRef]
- Mu, Q.; Zhao, M.; Running, S.W. Improvements to a modis global terrestrial evapotranspiration algorithm. Remote Sens. Environ. 2011, 115, 1781–1800. [Google Scholar] [CrossRef]
- Mu, Q.; Heinsch, F.A.; Zhao, M.; Running, S.W. Development of a global evapotranspiration algorithm based on modis and global meteorology data. Remote Sens. Environ. 2007, 111, 519–536. [Google Scholar] [CrossRef]
- Monteith, J. Evaporation and environment. In The State and Movement of Water in Living Organisms. Symposium of the Society of Experimental Biology; Cambridge University Press: Cambridge, UK, 1965; Volume 19, pp. 205–234. [Google Scholar]
- Tasumi, M.; Hirakawa, K.; Hasegawa, N.; Nishiwaki, A.; Kimura, R. Application of modis land products to assessment of land degradation of alpine rangeland in northern india with limited ground-based information. Remote Sens. 2014, 6, 9260–9276. [Google Scholar] [CrossRef]
- Mu, Q.; Zhao, M.; Running, S.W. Modis Global Terrestrial Evapotranspiration (et) Product (Nasa Mod16a2/a3); Algorithm Theoretical Basis Document, Collection; NASA: Washington, DC, USA, 2013; Volume 5. [Google Scholar]
- Wang, X.; Fan, J.; Xu, J.; Liu, F.; Gao, S.; Wei, X. Land surface emissivity change in china from 2001 to 2010. J. Geogr. Sci. 2012, 22, 407–415. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.S.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Stein, A.; Gerstner, K.; Kreft, H. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol. Lett. 2014, 17, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.T.; Packer, L. Habitat heterogeneity as a determinant of mammal species richness in high-energy regions. Nature 1997, 385, 252–254. [Google Scholar] [CrossRef]
- Olson, D.M.; Dinerstein, E.; Wikramanayake, E.D.; Burgess, N.D.; Powell, G.V.N.; Underwood, E.C.; D’Amico, J.A.; Itoua, I.; Strand, H.E.; Morrison, J.C. Terrestrial ecoregions of the world: A new map of life on earth. Bioscience 2009, 51, 933–938. [Google Scholar] [CrossRef]
- Olson, D.M.; Dinerstein, E. The Global 200: Priority ecoregions for global conservation. Ann. Mo. Bot. Gard. 2002, 89, 199–224. [Google Scholar] [CrossRef]
- Nagendra, H.; Rocchini, D.; Ghate, R.; Sharma, B.; Pareeth, S. Assessing plant diversity in a dry tropical forest: Comparing the utility of landsat and ikonos satellite images. Remote Sens. 2010, 2, 478–496. [Google Scholar] [CrossRef]
- Cleveland, W.S.; Devlin, S.J. Locally weighted regression: An approach to regression analysis by local fitting. J. Am. Stat. Assoc. 1988, 83, 596–610. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Loehlin, J.C. Latent Variable Models: An Introduction to Factor, Path, and Structural Analysis; Lawrence Erlbaum Associates Publishers: Mahwah, NJ, USA, 1998. [Google Scholar]
- Yao, Y.; Liang, S.; Li, X.; Chen, J.; Liu, S.; Jia, K.; Zhang, X.; Xiao, Z.; Fisher, J.B.; Mu, Q. Improving global terrestrial evapotranspiration estimation using support vector machine by integrating three process-based algorithms. Agric. For. Meteorol. 2017, 242, 55–74. [Google Scholar] [CrossRef]
- Eitrich, T.; Lang, B. Parallel Tuning of Support Vector Machine Learning Parameters for Large and Unbalanced Data Sets. In Proceedings of the International Symposium on Computational Life Science, Konstanz, Germany, 25–27 September 2005; Springer: Berlin, Germany, 2005; pp. 253–264. [Google Scholar]
- Vapnik, V. The nature of statistical learning theory. Technometrics 1995, 38, 409. [Google Scholar]
- Abdel-Rahman, E.M.; Mutanga, O.; Adam, E.; Ismail, R. Detecting sirex noctilio grey-attacked and lightning-struck pine trees using airborne hyperspectral data, random forest and support vector machines classifiers. ISPRS J. Photogramm. Remote Sens. 2014, 88, 48–59. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F.; Chang, C.-C.; Lin, C.-C.; Meyer, M.D. Package ‘e1071’; R Package Version; 2017. Available online: https://cran.r-project.org/web/packages/e1071/index.html (accessed on 12 November 2017).
- Sor, R.; Park, Y.-S.; Boets, P.; Goethals, P.L.; Lek, S. Effects of species prevalence on the performance of predictive models. Ecol. Model. 2017, 354, 11–19. [Google Scholar] [CrossRef]
- Cassano, A.; Marchese Robinson, R.L.; Palczewska, A.; Puzyn, T.; Gajewicz, A.; Tran, L.; Manganelli, S.; Cronin, M. Comparing the coral and random forest approaches for modelling the in vitro cytotoxicity of silica nanomaterials. Altern. Lab. Anim. 2016, 44, 533–556. [Google Scholar] [PubMed]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Goethals, P.L.; Dedecker, A.P.; Gabriels, W.; Lek, S.; De Pauw, N. Applications of artificial neural networks predicting macroinvertebrates in freshwaters. Aquat. Ecol. 2007, 41, 491–508. [Google Scholar] [CrossRef]
- Lek, S.; Guégan, J.-F. Artificial neural networks as a tool in ecological modelling, an introduction. Ecol. Model. 1999, 120, 65–73. [Google Scholar] [CrossRef]
- Lek, S.; Delacoste, M.; Baran, P.; Dimopoulos, I.; Lauga, J.; Aulagnier, S. Application of neural networks to modelling nonlinear relationships in ecology. Ecol. Model. 1996, 90, 39–52. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Kim, J.T.; Yun, G.Y. The neural network predictive model for heat island intensity in Seoul. Energy Build. 2016, 110, 353–361. [Google Scholar] [CrossRef]
- Van Echelpoel, W.; Boets, P.; Landuyt, D.; Gobeyn, S.; Everaert, G.; Bennetsen, E.; Mouton, A.; Goethals, P.L. Species distribution models for sustainable ecosystem management. In Developments in Environmental Modelling; Elsevier: New York, NY, USA, 2015; Volume 27, pp. 115–134. [Google Scholar]
- Ripley, B.; Venables, W.; Ripley, M.B. Package ‘nnet’; R Package Version 7.3-12; 2016. Available online: https://cran.r-project.org/web/packages/nnet/index.html (accessed on 12 November 2017).
- Myers, N. The biodiversity challenge: Expanded hot-spots analysis. Environmentalist 1990, 10, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.S.; Andelman, S.J.; Bakarr, M.I.; Boitani, L.; Brooks, T.M.; Cowling, R.M.; Fishpool, L.D.; da Fonseca, G.A.; Gaston, K.J.; Hoffmann, M. Effectiveness of the global protected area network in representing species diversity. Nature 2004, 428, 640. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, B.A.; Field, R.; Cornell, H.V.; Currie, D.J.; Guegan, J.; Kaufman, D.M.; Kerr, J.T.; Mittelbach, G.G.; Oberdorff, T.; Obrien, E.M. Energy, water, and broad-scale geographic patterns of species richness. Ecology 2003, 84, 3105–3117. [Google Scholar] [CrossRef]
- Gillman, L.N.; Wright, S.D. The influence of productivity on the species richness of plants: A critical assessment. Ecology 2006, 87, 1234–1243. [Google Scholar] [CrossRef]
- Isbell, F.; Craven, D.; Connolly, J.; Loreau, M.; Schmid, B.; Beierkuhnlein, C.; Bezemer, T.M.; Bonin, C.L.; Bruelheide, H.; De Luca, E. Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 2015, 526, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.H.; Currie, D.J.; Maurer, B.A. Energy supply and patterns of species richness on local and regional scales. In Species Diversity in Ecological Communities: Historical and Geographical Perspectives; University of Chicago Press: Chicago, IL, USA, 1993; pp. 66–74. [Google Scholar]
- Waide, R.B.; Willig, M.R.; Steiner, C.F.; Mittelbach, G.G.; Gough, L.; Dodson, S.I.; Juday, G.P.; Parmenter, R. The relationship between productivity and species richness. Annu. Rev. Ecol. Evol. System. 2003, 30, 257–300. [Google Scholar] [CrossRef]
- Pavlik, B.M. Species diversity in ecological communities: Historical and geographical perspectives. Madroño 1995, 42, 523–525. [Google Scholar]
- Rosenzweig, M.L. Species Diversity in Space and Time; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Currie, D.J.; Francis, A.P.; Kerr, J.T. Some general propositions about the study of spatial patterns of species richness. Ecoscience 1999, 6, 392–399. [Google Scholar] [CrossRef]
- Pacifici, M.; Visconti, P.; Butchart, S.H.; Watson, J.E.; Cassola, F.M.; Rondinini, C. Species/’traits influenced their response to recent climate change. Nat. Clim. Chang. 2017, 7, 205–208. [Google Scholar] [CrossRef]
- Davis, M.B. Lags in vegetation response to greenhouse warming. Clim. Chang. 1989, 15, 75–82. [Google Scholar] [CrossRef]
- Saatchi, S.; Asefi-Najafabady, S.; Malhi, Y.; Aragão, L.E.; Anderson, L.O.; Myneni, R.B.; Nemani, R. Persistent effects of a severe drought on amazonian forest canopy. Proc. Natl. Acad. Sci. USA 2013, 110, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; De Jeu, R.; Liu, Y.; Van der Werf, G.; Dolman, A. Using satellite based soil moisture to quantify the water driven variability in ndvi: A case study over mainland australia. Remote Sens. Environ. 2014, 140, 330–338. [Google Scholar] [CrossRef]
- Wu, D.; Zhao, X.; Liang, S.; Zhou, T.; Huang, K.; Tang, B.; Zhao, W. Time-lag effects of global vegetation responses to climate change. Glob. Chang. Biol. 2015, 21, 3520–3531. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, R.H.; MacArthur, J.W. On bird species diversity. Ecology 1961, 42, 594–598. [Google Scholar] [CrossRef]
- Stein, A.; Kreft, H. Terminology and quantification of environmental heterogeneity in species-richness research. Biol. Rev. 2015, 90, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Tamme, R.; Hiiesalu, I.; Laanisto, L.; Szava-Kovats, R.; Pärtel, M. Environmental heterogeneity, species diversity and co-existence at different spatial scales. J. Veg. Sci. 2010, 21, 796–801. [Google Scholar] [CrossRef]
- Gazol, A.; Tamme, R.; Price, J.N.; Hiiesalu, I.; Laanisto, L.; Pärtel, M. A negative heterogeneity–diversity relationship found in experimental grassland communities. Oecologia 2013, 173, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Laanisto, L.; Tamme, R.; Hiiesalu, I.; Szava-Kovats, R.; Gazol, A.; Pärtel, M. Microfragmentation concept explains non-positive environmental heterogeneity–diversity relationships. Oecologia 2013, 171, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Belmaker, J.; Jetz, W. Cross-scale variation in species richness–environment associations. Glob. Ecol. Biogeogr. 2011, 20, 464–474. [Google Scholar] [CrossRef]
- Pouteau, R.; Meyer, J.-Y.; Taputuarai, R.; Stoll, B. Support vector machines to map rare and endangered native plants in pacific islands forests. Ecol. Inform. 2012, 9, 37–46. [Google Scholar] [CrossRef]
- Lee, Y.; Lin, Y.; Wahba, G. Multicategory support vector machines: Theory and application to the classification of microarray data and satellite radiance data. J. Am. Stat. Assoc. 2004, 99, 67–81. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Liu, S.; Li, Y.; Xiao, Z.; Yao, Y.; Jiang, B.; Zhao, X.; Wang, X.; Xu, S. Global land surface fractional vegetation cover estimation using general regression neural networks from modis surface reflectance. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4787–4796. [Google Scholar] [CrossRef]
- Yang, L.; Jia, K.; Liang, S.; Liu, J.; Wang, X. Comparison of four machine learning methods for generating the glass fractional vegetation cover product from modis data. Remote Sens. 2016, 8, 682. [Google Scholar] [CrossRef]
- Sun, C.; Vilalta, R. Data selection using sash trees for support vector machines. In Proceedings of the Machine Learning and Data Mining in Pattern Recognition, Leipzig, Germany, 18–20 July 2007; pp. 286–295. [Google Scholar]
- Zhai, J.; Li, C.; Li, T. Sample selection based on kl divergence for effectively training svm. In Proceedings of the 2013 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Washington, DC, USA, 13–16 October 2013; pp. 4837–4842. [Google Scholar]
- Bellard, C.; Bertelsmeier, C.; Leadley, P.W.; Thuiller, W.; Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 2012, 15, 365–377. [Google Scholar] [CrossRef] [PubMed]








| Category | Metrics | Spatial Resolution | Temporal Resolution | Temporal Coverage |
|---|---|---|---|---|
| Dynamic habitat index (DHI)-cum | 1/12° | yearly | 2000–2011 | |
| Dynamic habitat index (DHI)-min | 1/12° | yearly | 2000–2011 | |
| Energy/productivity | Dynamic habitat index (DHI)-sea | 1/12° | yearly | 2000–2011 |
| Gross primary production (GPP) | 0.05° | monthly | 2001–2015 | |
| Potential Evapotranspiration (PET) | 0.05° | monthly | 2001–2014 | |
| Land surface temperature (LST) | 0.05° | monthly | 2000–2011 | |
| Climate | Evapotranspiration (ET) | 0.05° | monthly | 2000–2014 |
| Coefficient of variation (CV) | 1 Km | 5-year | 2001–2005 | |
| Evenness | 1 Km | 5-year | 2001–2005 | |
| Contrast (CON) | 1 Km | 5-year | 2001–2005 | |
| Dissimilarity (DIS) | 1 Km | 5-year | 2001–2005 | |
| Texture | Entropy (ENT) | 1 Km | 5-year | 2001–2005 |
| Homogeneity (HOM) | 1 Km | 5-year | 2001–2005 | |
| Range | 1 Km | 5-year | 2001–2005 | |
| Shannon | 1 Km | 5-year | 2001–2005 | |
| Simpson | 1 Km | 5-year | 2001–2005 | |
| Standard deviation (SD) | 1 Km | 5-year | 2001–2005 | |
| Correlation (COR) | 1 Km | 5-year | 2001–2005 | |
| Maximum (MAX) | 1 Km | 5-year | 2001–2005 | |
| Uniformity (UNI) | 1 Km | 5-year | 2001–2005 | |
| Variance (VAR) | 1 Km | 5-year | 2001–2005 |
| Species | SVM | RF | NN |
|---|---|---|---|
| Mammal | 4.514s | 13.635s | 2.639s |
| Bird | 4.465s | 14.252s | 2.651s |
| Amphibian | 3.497s | 14.173s | 2.421s |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Liang, S. Developing an Integrated Remote Sensing Based Biodiversity Index for Predicting Animal Species Richness. Remote Sens. 2018, 10, 739. https://doi.org/10.3390/rs10050739
Wu J, Liang S. Developing an Integrated Remote Sensing Based Biodiversity Index for Predicting Animal Species Richness. Remote Sensing. 2018; 10(5):739. https://doi.org/10.3390/rs10050739
Chicago/Turabian StyleWu, Jinhui, and Shunlin Liang. 2018. "Developing an Integrated Remote Sensing Based Biodiversity Index for Predicting Animal Species Richness" Remote Sensing 10, no. 5: 739. https://doi.org/10.3390/rs10050739
APA StyleWu, J., & Liang, S. (2018). Developing an Integrated Remote Sensing Based Biodiversity Index for Predicting Animal Species Richness. Remote Sensing, 10(5), 739. https://doi.org/10.3390/rs10050739