2.1. Bird Data
Bird data were obtained from the French Breeding Bird Survey (FBBS), a standardized monitoring program in which volunteer ornithologists identify and record breeding birds year after year [
30]. In the FBBS program, each observer provides a point location, and a randomly 2 km × 2 km survey square shaped plot is selected around a radius of 10 km from this location. Each plot contain 10 point counts with 100 m radii which are separated from each other by 300 m at a minimum and scattered in the landscape in order to best represent the diversity of habitats (
Figure 1). Each point count is performed twice a year during the breeding season (from April to mid-June) to record early singing species as well as last migrants. In each point count, heard or seen species are recorded during 5 min between 1 h and 4 h after sunrise. In our study, we focused the analyses on the year 2010 for which the sampling effort was high and well distributed at the national scale (a total of 1091 plots). For each point count in the surveyed plot squares, the maximum count over the two visits was retained for each species. We excluded raptors that have home range sizes (several tens of hectares) much higher than point count areas. We also excluded urban and wetland species because their habitats were poorly sampled compared to agricultural and woodland habitats. Moreover, the DHI components based on fPAR is is not available in the heart of the cities.
Plots were filtered according to the quality of the satellite images available on their locations (see
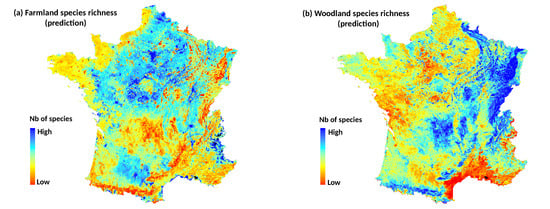
Section 2.2) and 759 plots were finally considered in the analyses. Because birds are expected to respond differently to landscape habitats according to their ecological requirements, four metrics of species richness (SR) per plot square were computed from the species observed at point counts. The first two metrics of SR were related to woodland species and farmland species (specialists of woodland and farmland habitats). The others were related to habitat generalists, and total species richness computed as the sum of all species (see
Appendix A for species names). These bird groups (guilds) were based on species information available at the national level (
http://vigienature.mnhn.fr).
2.2. Climate and Satellite Data
Because avifauna patterns are driven by large climate gradients at the French national scale, a map of macro-climate regions of France was used to derive a climate factor. The delineation of the climate regions in this map is based on 14 temperature and precipitation variables calculated from a 30-year period of climate data (1971–2000) provided by the french national meteorological service [
31]. The map is composed of eight regions (
Figure 2): the climate of mountain areas (type 1); the semi-continental climate and the climate of peripheral mountain areas (type 2); the degraded oceanic climate of the north and center plains (type 3); the altered oceanic climate (type 4); the marked oceanic climate (type 5); the altered Mediterranean climate (type 6); the southwest basin climate (type 7); and the marked Mediterranean climate (type 8). Based on this map, the dominant climate region was estimated for each plot of bird data and defined as one qualitative predictor. An alternative would have been to use WorldClim2 data which are locally defined at 1 km
scale [
32]. However, these data are based on an interpolation using fewer weather stations than the map defined by [
31] at French scale. This is why we retained the first source of data (i.e., map of macro-climate regions).
Normalized Difference Vegetation Index (NDVI) was used to provide surrogate of local vegetation cover and landscape heterogeneity patterns over France. The Terra-MODIS NDVI 16-day-composit product at 250-m spatial resolution (MOD13Q1) was retained. To accommodate for seasonal variation and evaluate the temporal effect of NDVI on models performance, five time periods in 2010 were selected (
Figure 2): late winter (6 March 2010, NDVI
), before leaves growing of deciduous tree; late spring (26 June 2010, NDVI
), before winter crops harvesting; late summer (29 August 2010, NDVI
), after winter crops harvesting before summer crops harvesting; end summer (14 September 2010, NDVI
), after partial summer crops harvesting; begin autumn (30 September 2010, NDVI
), after full crops harvesting. At these time periods, landscape composition and heterogeneity are revealed differently in NDVI data. In winter, NDVI values are low in the deciduous forests and in the non-cultivated agricultural parcels because of absence of photosynthetic activity. In spring and summer, NDVI reaches its highest values, mainly in winter and summer crops respectively, but also in grasslands and forests. In autumn, after all crops harvesting, NDVI discriminates well forests and open habitats because of higher difference of NDVI values, agricultural parcels being bare (see
Appendix E).
The NDVI data of each time period were selected after checking the MODIS quality layers of all the 16-day time series products of 2010. Based on the average value of the MOD13Q1
pixel reliability over France (0 = good, 1 = marginal data, 2 = snow/ice, 3 = cloudy), the images showing the highest quality (i.e., having the lowest average quality index) were retained for each time period. Then, a filter was applied on the bird survey plots available (
). Plots including NDVI pixels of bad quality were removed. Only plots composed of 100% of good pixels (i.e., with a
pixel reliability = 0) at the five time periods were used (
). Finally, NDVI values in these plots were summarized through several statistical predictors:
minimum,
maximum,
range,
average,
median, and
standard deviation. In order to prevent multicollinearity between covariates, only the
average (Avg) and
standard deviation (SD) predictors were retained (Spearman’s |
Rho|
) [
33]. The
average predictor is a proxy of landscape productivity and the dominant land cover in the plot. The
standard deviation reflects landscape heterogeneity.
To examine whether seasonal variations effect of NDVI on the models performance depends on the landscape context, we used land cover/land use datasets to calculate landscape composition in each bird plot. The forest layer was obtained from a nationwide highly detailed vector spatial database (BDTopo®, French mapping agency IGN) at 1-m resolution. For agricultural areas, the graphic parcel register (RPG) of 2010 was used. The French RPG database is produced annually since 2002 in application of the EU directives related to the Common Agricultural Policy. The land use information, based on a 28 class nomenclature (including several types of crops, as well as artificial and natural grasslands), is obtained from the farmers’ declaration of their cultivated areas. The information is defined at the agricultural block level composed of one single field or several contiguous ones. For each block, the proportion of each crop type is provided but without the spatial distribution of crops within the block.
The Dynamic Habitat Index of 2010 was also calculated at the national scale, in order to compare the performance of the DHI-based model with the ones based on single date NDVI. The DHI was derived from the most detailed MODIS fPAR (fraction of Photosynthetically Active Radiation) 8-day composite product at 500-m spatial resolution (MOD15A2H) [
34]. It includes three components [
26]: (1) the cumulative annual fPAR (DHI cum) reflecting the overall potential of vegetation productivity; (2) the minimum annual fPAR value (DHI min) indicating the minimum amount of vegetated cover; and (3) the coefficient of variation of annual fPAR (DHI cv) expressing the seasonality in productivity (
Figure 3). The
average and
standard deviation of the three DHI components were calculated for each plot of bird data (
) and used as dependent variables. All the variables were retained in the bird-habitat models after checking correlations between them (Spearman’s |
Rho|
). In order to avoid bias in the comparison with NDVI-based models because of difference in spatial resolution, NDVI variables were also computed from the MODIS product at 500-m (MOD13A1). To evaluate the seasonal effect of NDVI on models’s performance independently of DHI (see above), we kept the most detailed NDVI data at 250-m. This spatial resolution is likely to produce models with higher explanatory and predictive performances [
21].
2.3. Statistical Analysis
In a first time, we built models at national scale using the total filtered bird dataset (). We compared explanatory and predictive performances of NDVI data at several time periods with DHI data. For each of the four species richness metrics, we first constructed a model with only the climate variable, i.e., the dominant climate region of the bird plot, as predictor (e.g., SR~Climate). Then, we added into the climate model both NDVI variables (Avg and SD) of each time period in five separate models (e.g., SR~Climate + Avg.NDVI + SD.NDVI). Finally, we fitted a final model including DHI variables (6 predictors related to the Avg and SD of the three DHI components, see above) and the climate predictor.
In a second time, we examined whether the best time period of NDVI depended on the plot land cover context. This analysis was only applied to woodland and farmland species richness that were the best explained bird metrics at national scale. We partitioned bird national data into subsets, according to the dominant land cover of the bird plot squares. Landscape composition in each plot was calculated as the proportion of area occupied by four land covers: woody vegetation (including hedgerows), permanent grasslands, winter crops (mainly wheat and barley) and summer crops (mainly maize and sunflower). Because agricultural blocks of the land cover map (RPG) may cover partially the bird plot squares, only the blocks included by at least 70% in the plots were conserved for the estimation. Based on the land cover proportions, four subsets of bird plots were defined with the constraint to obtain a sample size large enough (i.e., with
) and a high dominance of a specific land cover (
Table 1). For each subset, we built models with climate and NDVI variables of each date as for the national scale.
Relationships between bird species richness and predictors were explored using generalized additive models (GAMs) with R version 3.1.0 software [
35] and the
gam package [
36]. The general expression of GAM model is [
37]:
where
Y is the response variable,
the predictors,
the non-parametric smooth functions and
g the link function relating to the expected value of
Y.
GAMs present the advantage to consider non-linear effects between response and predictor variables. Poisson distribution and a
log link function were used to calibrate the models. The most parsimonious models (i.e., with the lowest Akaike Information Criterion values) were selected by stepwise backward and forward variable selection [
38]. In addition, the procedure was allowed to select from one to four degrees of smoothing (1 = no smoothing) for each quantitative variable in GAMs.
The goodness of fit of the models (explanatory performance) was quantified by the amount of explained deviance (%
). The independent contribution of each variable to explain species richness was also estimated from a hierarchical partitioning procedure [
39].
The predictive performance was calculated by
k-fold cross-validation [
40]. The initial dataset was splitted randomly in three groups of equal size (
). Two groups were used to calibrate the model and the third group was used as a test set to evaluate predictions. The concordance between predictions and observations was measured with the Spearman rank correlations (
Rho) ranging from
to +1 [
41]. The correlation is considered to be weak for
<
Rho < 0.25, fair for 0.25 <
Rho < 0.50, moderate for 0.50 <
Rho < 0.75 and strong for
Rho > 0.75 [
42]. Cross-validation was repeated 100 times per model to obtain a variance of
Rho. The predictive performances between the models were compared using
Wilcoxon rank-sum non-parametric tests.
In order to assess whether the combination of multiple NDVI time periods lead to higher predictive performances than the use of a single date, a consensus approach was also followed. Because NDVI variables of different time periods were often highly correlated (
Rho > 0.70) they could not be included simultaneously into a single model [
33]. Alternatively, the consensus approach provides means to combine ensembles of model predictions, and to test if the predictive performance is higher after combination. This approach is usually adopted in species distribution modelling to combine outputs of single modelling techniques and to reduce uncertainty of predictions [
43]. Here, the outputs of single-date models were combined with the Weighted Average (WA) consensus method [
43]. The predictions were calculated as the mean of predictions of single-date models weighted by their individual predictive performance (mean
Rho). The concordance between predictions and observations was evaluated in the same way than the models based on single date.
Spatial autocorrelation of residuals was tested for each best model by computing non parametric spline correlograms using the
ncf package version 1.1-3 for R [
44]. Neighbours were considered until a distance of 300 km. Because a positive inherent autocorrelation was observed only for the very close surveyed plot squares (Spatial autocorrelation < 0.2 within 20 km; average distance of the closest square = 8.15 ± 7.01 km;
Appendix B), we considered the potential effect as not significant in the analysis [
45].