Semiautomated Detection and Mapping of Vegetation Distribution in the Antarctic Environment Using Spatial-Spectral Characteristics of WorldView-2 Imagery
Abstract
:1. Introduction
2. Study Area and Geospatial Data
2.1. Study Area
2.2. Geospatial Data
2.2.1. Satellite Data
2.2.2. Ground Reference and Supplementary Data
3. Geospatial Analysis
3.1. Data Pre-Processing
3.2. Feature Identification and Mapping
3.2.1. Customized Normalized Difference Vegetation Index (NDVI) Approach
3.2.2. Spectral Processing or Matching–Based Extraction Approach
3.2.3. Target Detection Approach
3.2.4. Pixel-Wise Supervised Classification Approach
3.3. Accuracy Assessment
4. Results
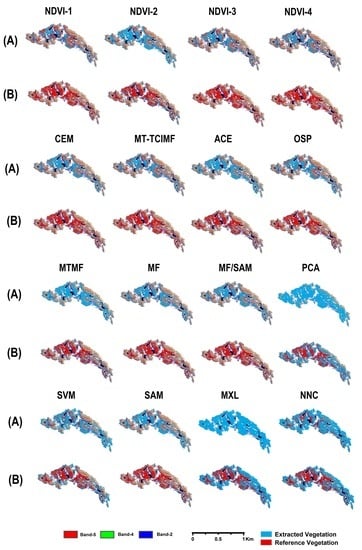
4.1. Performance of the Customized Normalized Difference Vegetation Index (NDVI) Approach
4.2. Performance of the Target Detection Approach
4.3. Performance of the Spectral Processing Approach
4.4. Performance of the Pixel-Wise Supervised Classification Approach
4.5. Overall Performance of Semi-Automatic Extraction Methods
4.6. Cross-Validation of Results
5. Discussion
5.1. Performance of Semi-Automatic Mapping Methods
5.2. Effect of Spectral-Spatial Resolution nn Sparse Vegetation Mapping
5.3. Comparison of Results with Previous Case Studies Dealing with Vegetation Mapping in Cryospheric Environments
5.4. Challenges in Vegetation Mapping in Cryospheric Environments
5.5. Experimental Limitations of the Present Study and a Way Forward
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turner, J.; Bindschadler, R.A.; Convey, P.; di Prisco, G.; Fahrbach, E.; Gutt, J.; Hodgson, D.A.; Mayewski, P.A.; Summerhayes, C.P. Antarctic Climate Change and the Environment (ACCE); Scientific Committee on Antarctic Research: Cambridge, UK, 2009; p. 526. [Google Scholar]
- Turner, J.; Barrand, N.; Bracegirdle, T.; Convey, P.; Hodgson, D.; Jarvis, M.; Jenkins, A.; Marshall, G.; Meredith, M.; Roscoe, H.; et al. Antarctic climate change and the environment: An update. Polar Record 2013, 50, 237–259. [Google Scholar] [CrossRef]
- Convey, P. Antarctic terrestrial biodiversity in a changing world. Polar Biol. 2011, 34, 1629–1641. [Google Scholar] [CrossRef]
- Smith, R.I.L. Terrestrial biology of the Antarctic and subAntarctic. In Antarctic Ecology, 2nd ed.; Laws, R.M., Ed.; Academic Press: London, UK, 1984; p. 61e162. [Google Scholar]
- Longton, R. The Biology of Polar Bryophytes and Lichens; CUP Archive: Cambridge, UK, 1988. [Google Scholar]
- Convey, P.; Chown, S.; Clarke, A.; Barnes, D.; Bokhorst, S.; Cummings, V.; Ducklow, H.; Frati, F.; Green, T.; Gordon, S.; et al. The spatial structure of Antarctic biodiversity. Ecol. Monogr. 2014, 84, 203–244. [Google Scholar] [CrossRef] [Green Version]
- Simms, É.; Ward, H. Multisensor NDVI-based monitoring of the tundra-taiga interface (Mealy Mountains, Labrador, Canada). Remote Sens. 2013, 5, 1066–1090. [Google Scholar] [CrossRef]
- Cannone, N.; Dalle Fratte, M.; Convey, P.; Worland, M.R.; Guglielmin, M. Ecology of moss banks at Signy Island (maritime Antarctica). Bot. J. Linnean Soc. 2017, 184, 518–533. [Google Scholar] [CrossRef]
- Cannone, N.; Guglielmin, M.; Convey, P.; Worland, M.R.; Favero Longo, S.E. Vascular plant changes in extreme environments: Effects of multiple drivers. Clim. Chang. 2016, 134, 651–665. [Google Scholar] [CrossRef]
- Convey, P.; Bindschadler, R.; di Prisco, G.; Fahrbach, E.; Gutt, J.; Hodgson, D.; Mayewski, P.; Summerhayes, C.; Turner, J. Antarctic climate change and the environment. Antarctic Sci. 2009, 21, 541. [Google Scholar] [CrossRef]
- Fowbert, J.A.; Smith, R.I.L. Rapid population increase in native vascular plants in the Argentine Islands, Antarctic Peninsula. Arctic Alpine Res. 1994, 26, 290–296. [Google Scholar] [CrossRef]
- Parnikoza, I.; Convey, P.; Dykyy, I.; Trakhimets, V.; Milinevsky, G.; Tyschenko, O.; Inozemtseva, D.; Kozeretska, I. Current status of the Antarctic herb tundra formation in the central Argentine Islands. Glob. Chang. Biol. 2009, 15, 1685–1693. [Google Scholar] [CrossRef]
- Robinson, S.; Wasley, J.; Tobin, A. Living on the edge - plants and global change in continental and maritime Antarctica. Global Chang. Biol. 2003, 9, 1681–1717. [Google Scholar] [CrossRef] [Green Version]
- Turner, D.; Lucieer, A.; Malenovský, Z.; King, D.; Robinson, S. Spatial co-registration of ultra-high resolution visible, multispectral and thermal images acquired with a micro-UAV over Antarctic Moss Beds. Remote Sens. 2014, 6, 4003–4024. [Google Scholar] [CrossRef]
- Lucieer, A.; Turner, D.; King, D.; Robinson, S. Using an Unmanned Aerial Vehicle (UAV) to capture micro-topography of Antarctic moss beds. Int. J. Appl. Earth Obs. Geoinformat. 2014, 27, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Bricher, P.; Lucieer, A.; Shaw, J.; Terauds, A.; Bergstrom, D. Mapping sub-antarctic cushion plants using random forests to combine very high resolution satellite imagery and terrain modelling. PLoS ONE 2013, 8, e72093. [Google Scholar] [CrossRef] [PubMed]
- Laidler, G.; Treitz, P.; Atkinson, D. Remote sensing of arctic vegetation: Relations between the NDVI, spatial resolution and vegetation cover on Boothia Peninsula, Nunavut. ARCTIC 2008, 61, 1. [Google Scholar] [CrossRef]
- Bhatt, U.; Walker, D.; Raynolds, M.; Comiso, J.; Epstein, H.; Jia, G.; Gens, R.; Pinzon, J.; Tucker, C.; Tweedie, C.; et al. Circumpolar arctic tundra vegetation change is linked to sea ice decline. Earth Interact. 2010, 14, 1–20. [Google Scholar] [CrossRef]
- Fretwell, P.; Convey, P.; Fleming, A.; Peat, H.; Hughes, K. Detecting and mapping vegetation distribution on the Antarctic Peninsula from remote sensing data. Polar Biol. 2011, 34, 273–281. [Google Scholar] [CrossRef]
- Malenovský, Z.; Lucieer, A.; King, D.; Turnbull, J.; Robinson, S. Unmanned aircraft system advances health mapping of fragile polar vegetation. Methods Ecol. Evolut. 2017, 8, 1842–1857. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.; Lucieer, A.; Williams, R. Texture-based classification of sub-Antarctic vegetation communities on Heard Island. Int. J. Appl. Earth Obs. Geoinformat. 2010, 12, 138–149. [Google Scholar] [CrossRef]
- Casanovas, P.; Black, M.; Fretwell, P.; Convey, P. Mapping lichen distribution on the Antarctic Peninsula using remote sensing, lichen spectra and photographic documentation by citizen scientists. Polar Res. 2015, 34, 25633. [Google Scholar] [CrossRef]
- Shin, J.; Kim, H.; Kim, S.; Hong, S. Vegetation abundance on the Barton Peninsula, Antarctica: Estimation from high-resolution satellite images. Polar Biol. 2014, 37, 1579–1588. [Google Scholar] [CrossRef]
- Markon, C.; Derksen, D. Identification of tundra land cover near Teshekpuk Lake, Alaska Using SPOT Satellite Data. ARCTIC 1994, 47, 222–231. [Google Scholar] [CrossRef]
- Griffith, B.; Douglas, C.D.; Noreen, E.; Donald, W.; Young, D.; Mccabe, T.R.; Russell, D.E.; White, R.G.; Cameron, R.D.; Whitten, R.K. The Porcupine Caribou Herd. In Biological Science Report; USGS/BRD: Reston, VA, USA, 2002. [Google Scholar]
- Walker, D.; Raynolds, M.; Daniëls, F.; Einarsson, E.; Elvebakk, A.; Gould, W.; Katenin, A.; Kholod, S.; Markon, C.; Melnikov, E.; et al. The other members of the CAVM Team. The Circumpolar Arctic vegetation map. J. Vegetat. Sci. 2005, 16, 267–282. [Google Scholar] [CrossRef]
- Lindblad, K.; Nyberg, R.; Molau, U. Generalization of heterogeneous alpine vegetation in air photo-based image classification, Latnjajaure catchment, northern Sweden. Pirineos 2006, 161, 3–32. [Google Scholar] [CrossRef]
- Karlsen, R.; Malnes, E.; Haarpaintner, J.; Solberg, R. Mapping and modelling the snowmelt and greening pattern in southern Norway by combining microwave and optical remote sensing sensors. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium IGARSS, Barcelona, Spain, 23–28 July 2007. [Google Scholar]
- Schneider, J.; Grosse, G.; Wagner, D. Land cover classification of tundra environments in the Arctic Lena Delta based on Landsat 7 ETM+ data and its application for upscaling of methane emissions. Remote Sens. Environ. 2009, 113, 380–391. [Google Scholar] [CrossRef] [Green Version]
- Raynolds, M.; Walker, D.; Epstein, H.; Pinzon, J.; Tucker, C. A new estimate of tundra-biome phytomass from trans-Arctic field data and AVHRR NDVI. Remote Sens. Lett. 2011, 3, 403–411. [Google Scholar] [CrossRef]
- Atkinson, D.; Treitz, P. Arctic Ecological Classifications Derived from Vegetation Community and Satellite Spectral Data. Remote Sens. 2012, 4, 3948–3971. [Google Scholar] [CrossRef] [Green Version]
- Ullmann, T.; Schmitt, A.; Roth, A.; Duffe, J.; Dech, S.; Hubberten, H.; Baumhauer, R. Land Cover Characterization and Classification of Arctic Tundra Environments by Means of Polarized Synthetic Aperture X- and C-Band Radar (PolSAR) and Landsat 8 Multispectral Imagery—Richards Island, Canada. Remote Sens. 2014, 6, 8565–8593. [Google Scholar] [CrossRef]
- Bratsch, S.; Epstein, H.; Buchhorn, M.; Walker, D. Differentiating among Four Arctic Tundra Plant Communities at Ivotuk, Alaska Using Field Spectroscopy. Remote Sens. 2016, 8, 51. [Google Scholar] [CrossRef]
- Black, M.; Casanovas, P.; Convey, P.; Fretwell, P. High Resolution mapping of Antarctic vegetation communities using airborne Hyperspectral data. In Proceedings of the RSPSOC, Aberystwyth, UK, 2–15 September 2014. [Google Scholar] [CrossRef]
- Buchhorn, M.; Walker, D.; Heim, B.; Raynolds, M.; Epstein, H.; Schwieder, M. Ground-based hyperspectral characterization of Alaska tundra vegetation along environmental gradients. Remote Sens. 2013, 5, 3971–4005. [Google Scholar] [CrossRef]
- Dalmayne, J.; Möckel, T.; Prentice, H.; Schmid, B.; Hall, K. Assessment of fine-scale plant species beta diversity using WorldView-2 satellite spectral dissimilarity. Ecol. Informat. 2013, 18, 1–9. [Google Scholar] [CrossRef]
- Liu, N.; Treitz, P. Modelling high arctic percent vegetation cover using field digital images and high resolution satellite data. Int. J. Appl. Earth Obs. Geoinformat. 2016, 52, 445–456. [Google Scholar] [CrossRef]
- Davidson, S.; Santos, M.; Sloan, V.; Watts, J.; Phoenix, G.; Oechel, W.; Zona, D. Mapping Arctic Tundra Vegetation Communities Using Field Spectroscopy and Multispectral Satellite Data in North Alaska, USA. Remote Sens. 2016, 8, 978. [Google Scholar] [CrossRef]
- Liu, N.; Budkewitsch, P.; Treitz, P. Examining spectral reflectance features related to Arctic percent vegetation cover: Implications for hyperspectral remote sensing of Arctic tundra. Remote Sens. Environ. 2017, 192, 58–72. [Google Scholar] [CrossRef]
- Juutinen, S.; Virtanen, T.; Kondratyev, V.; Laurila, T.; Linkosalmi, M.; Mikola, J.; Nyman, J.; Räsänen, A.; Tuovinen, J.; Aurela, M. Spatial variation and seasonal dynamics of leaf-area index in the arctic tundra-implications for linking ground observations and satellite images. Environ. Res. Lett. 2017, 12, 095002. [Google Scholar] [CrossRef]
- Gould, W.; Edlund, S.; Zoltai, S.; Raynolds, M.; Walker, D.; Maier, H. Canadian Arctic vegetation mapping. Int. J. Remote Sens. 2002, 23, 4597–4609. [Google Scholar] [CrossRef]
- Guay, K.; Beck, P.; Berner, L.; Goetz, S.; Baccini, A.; Buermann, W. Vegetation productivity patterns at high northern latitudes: A multi-sensor satellite data assessment. Global Chang. Biol. 2014, 20, 3147–3158. [Google Scholar] [CrossRef]
- Stow, D.; Hope, A.; McGuire, D.; Verbyla, D.; Gamon, J.; Huemmrich, F.; Houston, S.; Racine, C.; Sturm, M.; Tape, K.; et al. Remote sensing of vegetation and land-cover change in Arctic Tundra Ecosystems. Remote Sens. Environ. 2004, 89, 281–308. [Google Scholar] [CrossRef] [Green Version]
- McFadden, J.; Chapin, F.; Hollinger, D. Subgrid-scale variability in the surface energy balance of arctic tundra. J. Geophys. Res. Atmos. 1998, 103, 28947–28961. [Google Scholar] [CrossRef]
- Raynolds, M.; Walker, D.; Maier, H. NDVI patterns and phytomass distribution in the circumpolar Arctic. Remote Sens. Environ. 2006, 102, 271–281. [Google Scholar] [CrossRef]
- Hodgson, D.; Noon, P.; Vyverman, W.; Bryant, C.; Gore, D.; Appleby, P.; Gilmour, M.; Verleyen, E.; Sabbe, K.; Jones, V.; et al. Were the Larsemann Hills ice-free through the Last Glacial Maximum? Antarctic Sci. 2001, 13, 440–454. [Google Scholar] [CrossRef] [Green Version]
- Hodgson, D.; Verleyen, E.; Sabbe, K.; Squier, A.; Keely, B.; Leng, M.; Saunders, K.; Vyverman, W. Late Quaternary climate-driven environmental change in the Larsemann Hills, East Antarctica, multi-proxy evidence from a lake sediment core. Quat. Res. 2005, 64, 83–99. [Google Scholar] [CrossRef]
- Antarctic Treaty Consultative Meeting (ATCM). XXXVII—CEP XVII Final Report, Management Plan for Antarctic Specially Protected Area No. 174: Stornes, Larsemann Hills, Princess Elizabeth Land (Measure 12) and Larsemann Hills, East Antarctica Antarctic Specially Managed Area Management Plan (Measure 15), 28 April–7 May 2014, Brasilia, Brazil. Available online: https://www.ats.aq/documents/recatt/att555_e.pdf (accessed on 6 July 2017).
- Jawak, S.; Luis, A. Synergistic use of multitemporal RAMP, ICESat and GPS to construct an accurate DEM of the Larsemann Hills region, Antarctica. Adv. Space Res. 2012, 50, 457–470. [Google Scholar] [CrossRef]
- Harris, U. Larsemann Hills—Mapping from Aerial Photography Captured February 1998, Australian Antarctic Data Centre—CAASM Metadata, 2002, (Updated 2014). Available online: http://data.aad.gov.au/aadc/metadata/ (accessed on 6 July 2015).
- Paul, F.; Kääb, A. Perspectives on the production of a glacier inventory from multispectral satellite data in Arctic Canada: Cumberland Peninsula, Baffin Island. Ann. Glacio. 2005, 42, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Congalton, R. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Congalton, R.; Green, K. Assessing the accuracy of remotely sensed data. In Mapping Science; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Santos, T.; Freire, S. Testing the Contribution of WorldView-2 Improved Spectral Resolution for Extracting Vegetation Cover in Urban Environments. Can. J. Remote Sens. 2015, 41, 505–514. [Google Scholar] [CrossRef]
- Jawak, S.; Luis, A. A spectral index ratio-based Antarctic land-cover mapping using hyperspatial 8-band WorldView-2 imagery. Polar Sci. 2013, 7, 18–38. [Google Scholar] [CrossRef] [Green Version]
- DigitalGlobe. The Benefits of the Eight Spectral Bands of WorldView-2, White Paper (WP-8SPEC) 2010, Rev 01/13. Available online: http://www.geoimage.com.au/CaseStudies/TheBenefits_8BandData.pdf (accessed on 6 July 2014).
- Digital Globe. Radiometric Use of WorldView-2 Imagery. 2012. Available online: www.digitalglobe.com/downloads/Radiometric_Use_of_WorldView-2_Imagery.pdf (accessed on 6 July 2014).
- Richter, R.; Schläpfer, D. Atmospheric/Topographic Correction for Satellite Imagery; DLR Report DLR-IB 565-01/05; DLR: Wessling, Germany, 2011. [Google Scholar]
- Johnson, B. Effects of Pansharpening on Vegetation Indices. ISPRS Int. J. Geo-Inf. 2014, 3, 507–522. [Google Scholar] [CrossRef]
- Jawak, S.; Luis, A. A Comprehensive Evaluation of PAN-Sharpening Algorithms Coupled with Resampling Methods for Image Synthesis of Very High Resolution Remotely Sensed Satellite Data. Adv. Remote Sens. 2013, 2, 332–344. [Google Scholar] [CrossRef] [Green Version]
- Tucker, C. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.; Huete, A. Interpreting vegetation indices. Prevent. Vet. Med. 1991, 11, 185–200. [Google Scholar] [CrossRef]
- Myneni, R.; Hall, F.; Sellers, P.; Marshak, A. The interpretation of spectral vegetation indexes. IEEE Trans. Geosci. Remote Sens. 1995, 33, 481–486. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. In Proceedings of the 3rd Earth Resource Technology Satellite (ERTS) Symposium, NASA, Washington, DC, USA, 1 January 1974; pp. 309–317. [Google Scholar]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Kim, M.S.; Daughtry, C.S.T.; Chappelle, E.W.; McMurtrey, J.E.; Walthall, C.L. The use of high spectral resolution bands for estimating absorbed photosynthetically active radiation. In Proceedings of the 6th Symposium on Physical Measurements and Signatures in Remote Sensing, Val d’Isere, France, 1 January 1994; pp. 299–306. [Google Scholar]
- Daughtry, C. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Roujean, J.; Breon, F. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Broge, N.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Signature Analysis of Leaf Reflectance Spectra: Algorithm Development for Remote Sensing of Chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Xiaocheng, Z.; Tamas, J.; Chongcheng, C.; Malgorzata, W.V. Urban land cover mapping based on object oriented classification using worldview 2 satellite remote sensing images. In Proceedings of the International Scientific Conference on Sustainable Development & Ecological Footprint, Sopron, Hungary, 25–26 March 2012. [Google Scholar]
- Jawak, S.; Luis, A. A semiautomatic extraction of antarctic lake features using worldview-2 imagery. Photogramm. Eng. Remote Sens. 2014, 80, 939–952. [Google Scholar] [CrossRef]
- Wolf, A. Using WorldView-2 Vis-NIR multispectral imagery to support land mapping and feature extraction using normalized difference index ratios. In Proceedings of the SPIE 8390, Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XVIII, Baltimore, MD, USA, 23–27 April 2012. [Google Scholar]
- Peng, H.; Fuhui Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Jin, X.; Paswaters, S.; Cline, H. A comparative study of target detection algorithms for hyperspectral imagery. In Proceedings of the SPIE 7334, Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XV, Orlando, FL, USA, 27 April 2009. [Google Scholar]
- Jawak, S.; Luis, A. Very high-resolution satellite data for improved land cover extraction of Larsemann Hills, Eastern Antarctica. J. Appl. Remote Sens. 2013, 7, 073460. [Google Scholar] [CrossRef]
- Harsanyi, J.; Chang, C. Hyperspectral image classification and dimensionality reduction: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote Sens. 1994, 32, 779–785. [Google Scholar] [CrossRef]
- Bourennane, S.; Fossati, C.; Cailly, A. Improvement of Target-Detection Algorithms Based on Adaptive Three-Dimensional Filtering. IEEE Trans. Geosci. Remote Sens. 2011, 49, 1383–1395. [Google Scholar] [CrossRef]
- Chang, C.; Liu, J.; Chieu, B.; Ren, H.; Wang, C.; Lo, C.; Chung, P.; Yang, C.; Ma, D. Generalized constrained energy minimization approach to subpixel target detection for multispectral imagery. Opt. Eng. 2000, 39, 1275. [Google Scholar]
- Boardman, J.W. Leveraging the high dimensionality of AVIRIS data for improved sub-pixel target unmixing and rejection of false positives: Mixture tuned matched filtering. In Proceedings of the AVIRIS Proceedings, Seventh JPL Airborne Earth Science Workshop, Orlando, FL, USA, 8–12 April 1998; Volume 97, p. 21. [Google Scholar]
- Peterson, M.; Horner, T.; Moore, F. Evolving matched filter transform pairs for satellite image processing. In Proceedings of the SPIE Defense, Security, and Sensing, Orlando, FL, USA, 19 May 2011. [Google Scholar]
- Williams, P.; Hunt, E. Estimation of leafy spurge cover from hyperspectral imagery using mixture tuned matched filtering. Remote Sens. Environ. 2002, 82, 446–456. [Google Scholar] [CrossRef]
- Vapnik, V. Statistical Learning Theory; J. Wiley: New York, NY, USA, 1998. [Google Scholar]
- Kruse, F.; Lefkoff, A.; Boardman, J.; Heidebrecht, K.; Shapiro, A.; Barloon, P.; Goetz, A. The spectral image processing system (SIPS)—Interactive visualization and analysis of imaging spectrometer data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Canty, M. Boosting a fast-neural network for supervised land cover classification. Comput. Geosci. 2009, 35, 1280–1295. [Google Scholar] [CrossRef]
- Tso, B.; Mather, P. Classification of multisource remote sensing imagery using a genetic algorithm and Markov random fields. IEEE Trans. Geosci. Remote Sens. 1999, 37, 1255–1260. [Google Scholar] [CrossRef]
- Lu, D.; Hetrick, S.; Moran, E. Impervious surface mapping with Quickbird imagery. Int. J. Remote Sens. 2011, 32, 2519–2533. [Google Scholar] [CrossRef] [Green Version]
- Myint, S.; Gober, P.; Brazel, A.; Grossman-Clarke, S.; Weng, Q. Per-pixel vs. object-based classification of urban land cover extraction using high spatial resolution imagery. Remote Sens. Environ. 2011, 115, 1145–1161. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with random forest using very high spatial resolution 8-band worldview-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinformat. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Myint, S.W.; Wang, S.; Liu, H.; He, Z. Exploring the potential of WorldView-2 red-edge band-based vegetation indices for estimation of mangrove leaf area index with machine learning algorithms. Remote Sens. 2017, 9, 1060. [Google Scholar] [CrossRef]
- Barry, K.M.; Stone, C.; Mohammed, C.L. Crown-scale evaluation of spectral indices for defoliated and discoloured eucalypts. Int. J. Remote Sens. 2008, 29, 47–69. [Google Scholar] [CrossRef]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of sentinel-2 red-edge bands for empirical estimation of green LAI and chlorophyll content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, L.; Packalen, P.; Rautiainen, M. Comparison of Sentinel-2 and Landsat 8 in the estimation of boreal forest canopy cover and leaf area index. Remote Sens. Environ. 2017, 195, 259–274. [Google Scholar] [CrossRef]
- Verrelst, J.; Muñoz, J.; Alonso, L.; Delegido, J.; Rivera, J.P.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and-3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Clevers, J.; Kooistra, L.; Van Den Brande, M. Using Sentinel-2 data for retrieving LAI and leaf and canopy chlorophyll content of a potato crop. Remote Sens. 2017, 9, 405. [Google Scholar] [CrossRef]
- Zagajewski, B.; Tommervik, H.; Bjerke, J.W.; Raczko, E.; Bochenek, Z.; Klos, A.; Jarocinska, A.; Lavender, S.; Ziolkowski, D. Intraspecific differences in spectral reflectance curves as indicators of reduced vitality in high-Arctic plants. Remote Sens. 2017, 9, 1289. [Google Scholar] [CrossRef]
- Cho, M.A.; Debba, P.; Mutanga, O.; Dudeni-Tlhone, N.; Magadla, T.; Khuluse, S.A. Potential utility of the spectral red-edge region of SumbandilaSat imagery for assessing indigenous forest structure and health. Int. J. Appl. Earth Obs. Geoinf. 2012, 16, 85–93. [Google Scholar] [CrossRef]
- Cui, Z.; Kerekes, J.P. Potential of red edge spectral bands in future landsat satellites on agroecosystem canopy green leaf area index retrieval. Remote Sens. 2018, 10, 1458. [Google Scholar] [CrossRef]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Peterson, S.H.; Alonzo, M. The impact of spatial resolution on the classification of plant species and functional types within imaging spectrometer data. Remote Sens. Environ. 2015, 171, 45–57. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Investigating the robustness of the new Landsat-8 Operational Land Imager derived texture metrics in estimating plantation forest aboveground biomass in resource constrained areas. ISPRS J. Photogramm. Remote Sens. 2015, 108, 12–32. [Google Scholar] [CrossRef]
- Mikheeva, A.I.; Tutubalina, O.V.; Zimin, M.V.; Golubeva, E.I. A Subpixel Classification of Multispectral Satellite Imagery for Interpretation of Tundra-Taiga Ecotone Vegetation (Case Study on Tuliok River Valley, Khibiny, Russia). Izv. Atmos. Ocean. Phys. 2017, 53, 1164. [Google Scholar] [CrossRef]
- Mikola, J.; Virtanen, T.; Linkosalmi, M.; Vähä, E.; Nyman, J.; Postanogova, O.; Räsänen, A.; Kotze, D.J.; Laurila, T.; Juutinen, S.; et al. Spatial variation and linkages of soil and vegetation in the Siberian Arctic tundra—Coupling field observations with remote sensing data. Biogeosciences 2018, 15, 2781–2801. [Google Scholar] [CrossRef]
- Suchá, R.; Jakešová, L.; Kupková, L.; Červená, L. Classification of vegetation above the treeline in the Krkonoše Mts. National Park using remote sensing multispectral data. AUC Geograph. 2016, 51, 113–129. Available online: http://www.aucgeographica.cz/index.php/aucg/article/view/71 (accessed on 6 July 2018). [CrossRef]
- Kupková, L.; Červená, L.; Suchá, R.; Zagajewski, B.; Březina, S.; Albrechtová, J. Classification of Tundra Vegetation in the Krkonoše Mts. National Park Using APEX, AISA Dual and Sentinel-2A Data. Eur. J. Remote Sens. 2017, 50, 29–46. [Google Scholar] [CrossRef]
- Ulrich, M.; Grosse, G.; Chabrillat, S.; Schirrmeister, L. Spectral characterization of periglacial surfaces and geomorphological units in the Arctic Lena Delta using field spectrometry and remote sensing. Remote Sens. Environ. 2009, 113, 1220–1235. [Google Scholar] [CrossRef] [Green Version]











| Dataset Details | Source of Datasets | Temporal Range (DD/MM/YY) | Utilization in the Present Study | ||||
|---|---|---|---|---|---|---|---|
| VM | VSD | MD | DEA | SDR | |||
| WorldView-2 multispectral (2m) and PAN (0.5 m) | DigitalGlobe | 09/02/2011 | ✗ | ✓ | ✗ | ✗ | ✗ |
| WorldView-2 GS-sharpened image (0.5m) | Processed | 09/02/2011 | ✓ | ✓ | ✓ | ✓ | ✓ |
| Google Earth Images | 31/12/1999, 24/02/2006, 03/03/2006, 04/01/2011 | ✗ | ✓ | ✗ | ✗ | ✓ | |
| Larsemann Hills, ASMA map series | AADC | Produced in 2013-14 | ✗ | ✓ | ✗ | ✗ | ✓ |
| Land cover map (1:2500) | SOI during Indian Antarctic Expedition | 2007–2008 (Sep–Mar) | ✗ | ✓ | ✗ | ✗ | ✓ |
| Aerial Photographs | InSEA | 2010–2012 (Sep–Mar) | ✗ | ✓ | ✗ | ✗ | ✗ |
| DGPS surveying | InSEA | 2008–2011 (Sep–Mar) | ✗ | ✓ | ✓ | ✓ | ✗ |
| DEM | Jawak and Luis [49] | 2003–2011 | ✗ | ✓ | ✗ | ✗ | ✓ |
| NDVI | NDVI Model | Formula | Threshold Range |
|---|---|---|---|
| NDVI-1 | NDVI(7-5/7+5) | 0.53−0.65 | |
| NDVI-2 | NDVI(8-5/8+5) | 0.57−0.62 | |
| NDVI-3 | NDVI(7-6/7+6) | 0.54−0.63 | |
| NDVI-4 | NDVI(8-6/8+6) | 0.55−0.66 |
| Category/Approach | Acronym | Method Name |
|---|---|---|
| Target detection | MT-TCIMF | Mixture Tuned Target-Constrained Interference-Minimized Filter |
| CEM | Constrained Energy Minimization | |
| ACE | Adaptive Coherence Estimator | |
| OSP | Orthogonal Subspace Projection | |
| Spectral processing | MTMF | Mixture Tuned Matched Filtering |
| MF | Matched Filtering | |
| MF/SAM | Matched Filtering/Spectral Angle Mapper Ratio | |
| PCA | Principle Component Analysis | |
| Pixel-wise supervised | MXL | Maximum Likelihood Classifier |
| classification | SVM | Support Vector Machine |
| NNC | Neural Net Classifier | |
| SAM | Spectral Angle Mapper |
| Bias in Extracted Vegetated Area (m2) | ||||||
|---|---|---|---|---|---|---|
| Experiment | Cross-Validation | |||||
| Approach | Reference | FI | SPN | Total | Average | SO |
| Customized NDVI Approach | ||||||
| NDVI-1 | Present work | 16107.10 | 311774.20 | 327881.30 | 163940.70 | 167060.08 |
| NDVI-2 | Present work | 14950.85 | 167791.00 | 182741.80 | 91370.90 | 114117.37 |
| NDVI-3 | Present work | 15518.06 | 284756.50 | 300274.60 | 150137.30 | 161572.11 |
| NDVI-4 | Present work | 15093.41 | 170286.70 | 185380.10 | 92690.06 | 117345.58 |
| Average | 15417.36 | 233652.10 | 249069.50 | 124534.70 | 140023.78 | |
| RMSE | 15423.91 | 242611.20 | 257600.10 | 128800.10 | 142133.19 | |
| Target Detection Approach | ||||||
| MT-TCIMF | [75] | 15349.75 | 259172.20 | 274522.00 | 137261.00 | 153017.34 |
| OSP | [77] | 15727.75 | 290478.20 | 306206.00 | 153103.00 | 164800.33 |
| ACE | [78] | −15470.50 | −279690.00 * | −2951610.00 * | −147580.00 * | −160926.47 |
| CEM | [79] | 16382.61 | 437713.50 | 454096.10 | 227048.00 | 225813.57 |
| Average | 7997.40 | 176918.40 | 184915.80 | 92457.90 | 95676.19 | |
| RMSE | 15737.72 | 324564.20 | 340017.50 | 170008.70 | 178509.49 | |
| Spectral Processing Approach | ||||||
| MTMF | [80] | −14083.00 | −84882.90 | −98965.90 | −49483.00 | −80866.76 |
| MF/SAM | [81] | 14329.33 | 145633.00 | 159962.30 | 79981.15 | 106046.83 |
| MF | [82] | 14405.00 | 169909.60 | 184314.60 | 92157.32 | 115085.83 |
| PCA | [75] | −146687.00 ^* | −744066.00 | −890752.00 | −445376.00 | −473579.00 |
| Average | −33008.90 | −128351.00 | −161360.00 | −80680.20 | −83328.27 | |
| RMSE | 74377.72 | 390805.90 | 464433.50 | 232216.70 | 252639.65 | |
| Pixel-wise Supervised Classification Approach | ||||||
| SVM | [83] | −18217.40 | −502521.00 | −520739.00 | −260369.00 | −240017.71 |
| SAM | [84] | 15989.08 | 361368.90 * | 377358.00 * | 188679.00 | 218388.68 |
| NNC | [85] | −95117.40 ^ | −518722.00 | −613839.00 | −306920.00 | −340737.99 |
| MXL | [86] | −155180.00 ^ | −488940.00 | −644120.00 | −322060.00 | −390291.08 |
| Average | −63131.50 | −287204.00 | −350335.00 | −175168.00 | −188164.52 | |
| RMSE | 91809.25 | 472030.20 | 548921.10 | 274460.60 | 305667.96 | |
| Total Average | −18181.40 | −1246.14 | −19427.50 | −9713.77 | −8948.21 | |
| Total RMSE | 60096.91 | 367336.30 | 418025.90 | 209012.90 | 228761.44 | |
| Misclassified Vegetated Pixels (%) | ||||||
|---|---|---|---|---|---|---|
| Experiment | Cross-Validation | |||||
| Approach | Reference | FI | SPN | Total | Average | SO |
| Customized NDVI | ||||||
| NDVI-1 | Present work | 2.50 | 14.22 | 11.55 | 8.36 | 10.35 |
| NDVI-2 | Present work | 2.32 | 7.65 | 6.44 | 4.99 | 7.07 |
| NDVI-3 | Present work | 2.41 | 12.99 | 10.58 | 7.70 | 10.01 |
| NDVI-4 | Present work | 2.34 | 7.77 | 6.53 | 5.05 | 7.27 |
| Average | 2.39 | 10.65 | 8.78 | 6.52 | 8.68 | |
| RMSE | 2.39 | 11.06 | 9.08 | 6.70 | 8.81 | |
| Target Detection Approach | ||||||
| MT-TCIMF | [75] | 2.38 | 11.82 | 9.67 | 7.10 | 9.48 |
| OSP | [77] | 2.44 | 13.25 | 10.79 | 7.84 | 10.21 |
| ACE | [78] | 2.40 | 12.75 | 10.40 | 7.58 | 9.97 |
| CEM | [79] | 2.54 | 19.96 * | 16.00 * | 11.25 * | 13.99 |
| Average | 2.44 | 14.44 | 11.72 | 8.44 | 10.91 | |
| RMSE | 2.44 | 14.80 | 11.98 | 8.60 | 11.06 | |
| Spectral Processing Approach | ||||||
| MTMF | [80] | 2.18 | 3.87 | 3.49 | 3.03 | 5.01 |
| MF/SAM | [81] | 2.22 | 6.64 | 5.64 | 4.43 | 6.57 |
| MF | [82] | 2.23 | 7.75 | 6.50 | 4.99 | 7.13 |
| PCA | [75] | 22.75 *^ | 33.93 * | 31.39 * | 28.34 *^ | 29.34 |
| Average | 7.35 | 13.05 | 11.75 | 10.20 | 12.01 | |
| RMSE | 11.54 | 17.82 | 16.37 | 14.64 | 15.65 | |
| Pixel-wise Supervised Classification Approach | ||||||
| SVM | [83] | 2.83 | 22.92 | 18.35 | 12.87 | 14.87 |
| SAM | [84] | 2.48 | 16.48 * | 13.30 | 9.48 | 13.53 |
| NNC | [85] | 14.75 ^ | 23.65 | 21.63 | 19.20 | 21.11 |
| MXL | [86] | 24.07 ^ | 22.30 | 22.70 | 23.18^ | 24.18 |
| Average | 11.03 | 21.34 | 19.00 | 16.18 | 18.42 | |
| RMSE | 14.24 | 21.53 | 19.34 | 17.04 | 18.94 | |
| Total Average | 5.80 | 14.87 | 12.81 | 10.34 | 12.51 | |
| Total RMSE | 9.32 | 16.75 | 14.73 | 12.49 | 14.17 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jawak, S.D.; Luis, A.J.; Fretwell, P.T.; Convey, P.; Durairajan, U.A. Semiautomated Detection and Mapping of Vegetation Distribution in the Antarctic Environment Using Spatial-Spectral Characteristics of WorldView-2 Imagery. Remote Sens. 2019, 11, 1909. https://doi.org/10.3390/rs11161909
Jawak SD, Luis AJ, Fretwell PT, Convey P, Durairajan UA. Semiautomated Detection and Mapping of Vegetation Distribution in the Antarctic Environment Using Spatial-Spectral Characteristics of WorldView-2 Imagery. Remote Sensing. 2019; 11(16):1909. https://doi.org/10.3390/rs11161909
Chicago/Turabian StyleJawak, Shridhar D., Alvarinho J. Luis, Peter T. Fretwell, Peter Convey, and Udhayaraj A. Durairajan. 2019. "Semiautomated Detection and Mapping of Vegetation Distribution in the Antarctic Environment Using Spatial-Spectral Characteristics of WorldView-2 Imagery" Remote Sensing 11, no. 16: 1909. https://doi.org/10.3390/rs11161909
APA StyleJawak, S. D., Luis, A. J., Fretwell, P. T., Convey, P., & Durairajan, U. A. (2019). Semiautomated Detection and Mapping of Vegetation Distribution in the Antarctic Environment Using Spatial-Spectral Characteristics of WorldView-2 Imagery. Remote Sensing, 11(16), 1909. https://doi.org/10.3390/rs11161909