Development and Validation of a Model to Combine NDVI and Plant Height for High-Throughput Phenotyping of Herbage Yield in a Perennial Ryegrass Breeding Program
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
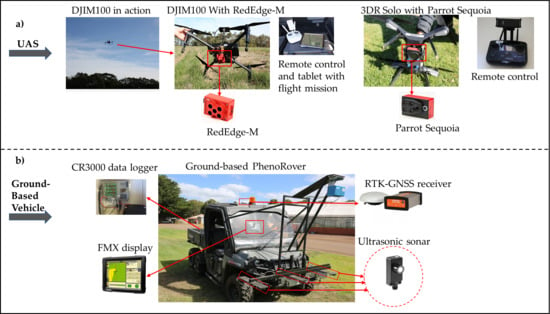
2.2. Data Collection Platforms
2.2.1. Unmanned Aerial Systems (UAS)
2.2.2. Image Processing and NDVI Data Extraction
2.2.3. PhenoRover System Field Deployments
2.2.4. Plant Height Data Extraction from the Ultrasonic Sonar
2.2.5. HY Data Collection
2.3. Estimation Models of HY through Sensors Data Combination
2.4. Statistical Analysis
3. Results
3.1. Seasonal Herbage Yield Variation
3.2. Correlation between Manual and Sonar Plant Height
3.3. Individual Plant Herbage Yield Model Evaluation
3.4. Herbage Yield Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foster, J.; Cogan, N. Pasture Molecular Genetic Technologies—Pre-Competitive Facilitated Adoption by Pasture Plant Breeding Companies; Meat & Livestock Australia Limited: Sydney, Australia, 2015. [Google Scholar]
- McDonagh, J.; O’Donovan, M.; McEvoy, M.; Gilliland, T.J. Genetic gain in perennial ryegrass (Lolium perenne) varieties 1973 to 2013. Euphytica 2016, 212, 187–199. [Google Scholar] [CrossRef]
- Hunt, C.L.; Jones, C.S.; Hickey, M.; Hatier, J.H.B. Estimation in the field of individual perennial ryegrass plant position and dry matter production using a custom-made high-throughput image analysis tool. Crop Sci. 2015, 55, 2910–2917. [Google Scholar] [CrossRef]
- Sampoux, J.-P.; Baudouin, P.; Bayle, B.; Béguier, V.; Bourdon, P.; Chosson, J.-F.; Deneufbourg, F.; Galbrun, C.; Ghesquière, M.; Noël, D.; et al. Breeding perennial grasses for forage usage: An experimental assessment of trait changes in diploid perennial ryegrass (Lolium perenne L.) cultivars released in the last four decades. Field Crops Res. 2011, 123, 117–129. [Google Scholar] [CrossRef]
- Gebremedhin, A.; Badenhorst, P.E.; Wang, J.; Spangenberg, G.C.; Smith, K.F. Prospects for measurement of dry matter yield in forage breeding programs using sensor technologies. Agronomy 2019, 9, 65. [Google Scholar] [CrossRef]
- Haghighattalab, A.; González Pérez, L.; Mondal, S.; Singh, D.; Schinstock, D.; Rutkoski, J.; Ortiz-Monasterio, I.; Singh, R.P.; Goodin, D.; Poland, J. Application of unmanned aerial systems for high throughput phenotyping of large wheat breeding nurseries. Plant Methods 2016, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Crain, J.; Mondal, S.; Rutkoski, J.; Singh, R.P.; Poland, J. Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Tanger, P.; Klassen, S.; Mojica, J.P.; Lovell, J.T.; Moyers, B.T.; Baraoidan, M.; Naredo, M.E.B.; McNally, K.L.; Poland, J.; Bush, D.R.; et al. Field-based high throughput phenotyping rapidly identifies genomic regions controlling yield components in rice. Sci. Rep. 2017, 7, 42839. [Google Scholar] [CrossRef] [Green Version]
- Slater, A.T.; Cogan, N.O.I.; Rodoni, B.C.; Daetwyler, H.D.; Hayes, B.J.; Caruana, B.; Badenhorst, P.E.; Spangenberg, G.C.; Forster, J.W. Breeding differently—The digital revolution: High-throughput phenotyping and genotyping. Potato Res. 2017, 60, 337–352. [Google Scholar] [CrossRef]
- Michez, A.; Lejeune, P.; Bauwens, S.; Herinaina, A.A.L.; Blaise, Y.; Castro Muñoz, E.; Lebeau, F.; Bindelle, J. Mapping and monitoring of biomass and grazing in pasture with an unmanned aerial system. Remote Sens. 2019, 11, 473. [Google Scholar] [CrossRef]
- Insua, J.R.; Utsumi, S.A.; Basso, B. Estimation of spatial and temporal variability of pasture growth and digestibility in grazing rotations coupling unmanned aerial vehicle (UAV) with crop simulation models. PLoS ONE 2019, 14, e0212773. [Google Scholar] [CrossRef]
- Schaefer, T.M.; Lamb, W.D. A combination of plant NDVI and LiDAR measurements improve the estimation of pasture biomass in tall fescue (Festuca arundinacea var. Fletcher). Remote Sens. 2016, 8, 109. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Observ. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Thorp, K.R.; Wang, G.; Bronson, K.F.; Badaruddin, M.; Mon, J. Hyperspectral data mining to identify relevant canopy spectral features for estimating durum wheat growth, nitrogen status, and grain yield. Comput. Electron. Agric. 2017, 136, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tilly, N.; Aasen, H.; Bareth, G. Fusion of plant height and vegetation indices for the estimation of barley biomass. Remote Sens. 2015, 7, 11449–11480. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Molero, G.; Stellacci, A.; Bort, J.; Nogues, S.; Araus, J. NDVI as a potential tool for predicting biomass, plant nitrogen content and growth in wheat genotypes subjected to different water and nitrogen conditions. Cereal Res. Commun. 2011, 39, 147–159. [Google Scholar] [CrossRef]
- Smith, R.C.G.; Wallace, J.F.; Hick, P.T.; Gilmour, R.F.; Belford, R.K.; Portmann, P.A.; Regan, K.L.; Turner, N.C. Potential of using field spectroscopy during early growth for ranking biomass in cereal breeding trials. Aust. J. Agric. Res. 1993, 44, 1713–1730. [Google Scholar] [CrossRef]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A direct comparison of remote sensing approaches for high-throughput phenotyping in plant breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef]
- Prabhakara, K.; Hively, W.D.; McCarty, G.W. Evaluating the relationship between biomass, percent groundcover and remote sensing indices across six winter cover crop fields in Maryland, United States. Int. J. Appl. Earth Observ. Geoinf. 2015, 39, 88–102. [Google Scholar] [CrossRef] [Green Version]
- Lu, N.; Zhou, J.; Han, Z.; Li, D.; Cao, Q.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cheng, T. Improved estimation of aboveground biomass in wheat from RGB imagery and point cloud data acquired with a low-cost unmanned aerial vehicle system. Plant Methods 2019, 15, 17. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, L.; Zhang, H.; Han, W.; Peng, X. Estimating above-ground biomass of maize using features derived from UAV-based RGB imagery. Remote Sens. 2019, 11, 1261. [Google Scholar] [CrossRef]
- Brocks, S.; Bareth, G. Estimating barley biomass with crop surface models from oblique RGB imagery. Remote Sens. 2018, 10, 268. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.T.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High throughput determination of plant height, ground cover, and above-ground biomass in wheat with LiDAR. Front. Plant Sci. 2018, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Li, J.; Bhatta, M.; Shi, Y.; Baenziger, S.P.; Ge, Y. Wheat height estimation using LiDAR in comparison to ultrasonic sensor and UAS. Sensors 2018, 18, 3731. [Google Scholar] [CrossRef] [PubMed]
- Barmeier, G.; Mistele, B.; Schmidhalter, U. Referencing laser and ultrasonic height measurements of barleycultivars by using a herbometre as standard. Crop Pasture Sci. 2016, 67, 1215–1222. [Google Scholar] [CrossRef]
- Pittman, J.J.; Arnall, B.D.; Interrante, M.S.; Moffet, A.C.; Butler, J.T. Estimation of biomass and canopy height in bermudagrass, alfalfa, and wheat using ultrasonic, laser, and spectral sensors. Sensors 2015, 15, 2920–2943. [Google Scholar] [CrossRef]
- Hu, P.; Chapman, S.C.; Wang, X.; Potgieter, A.; Duan, T.; Jordan, D.; Guo, Y.; Zheng, B. Estimation of plant height using a high throughput phenotyping platform based on unmanned aerial vehicle and self-calibration: Example for sorghum breeding. Eur. J. Agron. 2018, 95, 24–32. [Google Scholar] [CrossRef]
- Fricke, T.; Richter, F.; Wachendorf, M. Assessment of forage mass from grassland swards by height measurement using an ultrasonic sensor. Comput. Electron. Agric. 2011, 79, 142–152. [Google Scholar] [CrossRef]
- Fricke, T.; Wachendorf, M. Combining ultrasonic sward height and spectral signatures to assess the biomass of legume–grass swards. Comput. Electron. Agric. 2013, 99, 236–247. [Google Scholar] [CrossRef]
- Condorelli, G.E.; Maccaferri, M.; Newcomb, M.; Andrade-Sanchez, P.; White, J.W.; French, A.N.; Sciara, G.; Ward, R.; Tuberosa, R. Comparative aerial and ground based high throughput phenotyping for the genetic dissection of NDVI as a proxy for drought adaptive traits in durum wheat. Front. Plant Sci. 2018, 9, 893. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Trotter, M.; Robson, A.; Schneider, D.; Frizell, L.; Saint, A.; Lamb, D.; Blore, C. Estimating pasture biomass with active optical sensors. Adv. Anim. Biosci. 2017, 8, 754–757. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Li, C.; Li, Z.; Wang, Y.; Feng, H.; Xu, B. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Thompson, A.L.; Thorp, K.R.; Conley, M.; Andrade-Sanchez, P.; Heun, J.T.; Dyer, J.M.; White, J.W. Deploying a proximal sensing cart to identify drought-adaptive traits in upland cotton for high-throughput phenotyping. Front. Plant Sci. 2018, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Thorp, K.R.; White, J.W.; French, A.N.; Poland, J.A. Approaches for geospatial processing of field-based high-throughput plant phenomics data from ground vehicle platforms. Trans. ASABE 2016, 59, 1053–1067. [Google Scholar]
- Cullen, B.R.; Eckard, R.J.; Callow, M.N.; Johnson, I.R.; Chapman, D.F.; Rawnsley, R.P.; Garcia, S.C.; White, T.; Snow, V.O. Simulating pasture growth rates in Australian and New Zealand grazing systems. Aust. J. Agric. Res. 2008, 59, 761–768. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V. UAV-based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef]
- Patrick, A.; Li, C. High throughput phenotyping of blueberry bush morphological traits using unmanned aerial systems. Remote Sens. 2017, 9, 1250. [Google Scholar] [CrossRef]
- Mu, Y.; Fujii, Y.; Takata, D.; Zheng, B.; Noshita, K.; Honda, K.; Ninomiya, S.; Guo, W. Characterization of peach tree crown by using high-resolution images from an unmanned aerial vehicle. Hortic. Res. 2018, 5, 74. [Google Scholar] [CrossRef] [Green Version]
- Robertsen, D.C.; Hjortshøj, L.R.; Janss, L.L. Genomic selection in cereal breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef]
- Nakaya, A.; Isobe, S.N. Will genomic selection be a practical method for plant breeding? Ann. Bot. 2012, 110, 1303–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Wang, J.; Cogan, N.O.I.; Pembleton, L.W.; Badenhorst, P.; Forster, J.W.; Spangenberg, G.C.; Hayes, B.J.; Daetwyler, H.D. Optimizing resource allocation in a genomic breeding program for perennial ryegrass to balance genetic gain, cost, and inbreeding. Crop Sci. 2017, 57, 243–252. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Inch, C.; Baillie, R.C.; Drayton, M.C.; Thakur, P.; Ogaji, Y.O.; Spangenberg, G.C.; Forster, J.W.; Daetwyler, H.D.; Cogan, N.O.I. Exploitation of data from breeding programs supports rapid implementation of genomic selection for key agronomic traits in perennial ryegrass. Theor. Appl. Genet. 2018, 131, 1891–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grüner, E.; Astor, T.; Wachendorf, M. Biomass prediction of heterogeneous temperate grasslands using an SFM approach based on UAV imaging. Agronomy 2019, 9, 54. [Google Scholar] [CrossRef]
- Gonzalez, M.A.; Hussey, M.A.; Conrad, B.E. Plant height, disk, and capacitance meters used to estimate bermudagrass herbage mass. Agron. J. 1990, 82, 861–864. [Google Scholar] [CrossRef]
- Thompson, A.L.; Thorp, K.R.; Conley, M.M.; Elshikha, D.M.; French, A.N.; Andrade-Sanchez, P.; Pauli, D. Comparing nadir and multi-angle view sensor technologies for measuring in-field plant height of upland cotton. Remote Sens. 2019, 11, 700. [Google Scholar] [CrossRef]
- Goodwin, A.W.; Lindsey, L.E.; Harrison, S.K.; Paul, P.A. Estimating wheat yield with normalized difference vegetation index and fractional green canopy cover. Crop Forage Turfgrass Manag. 2018, 4. [Google Scholar] [CrossRef]
- Lamb, D.W.; Schneider, D.A.; Trotter, M.G.; Schaefer, M.T.; Yule, I.J. Extended-altitude, aerial mapping of crop NDVI using an active optical sensor: A case study using a Raptor™ sensor over wheat. Comput. Electron. Agric. 2011, 77, 69–73. [Google Scholar] [CrossRef]
- Zaman-Allah, M.; Vergara, O.; Araus, J.L.; Tarekegne, A.; Magorokosho, C.; Zarco-Tejada, P.J.; Hornero, A.; Albà, A.H.; Das, B.; Craufurd, P.; et al. Unmanned aerial platform-based multi-spectral imaging for field phenotyping of maize. Plant Methods 2015, 11, 35. [Google Scholar] [CrossRef]
- Park, C.-W.; Na, S.-I.; So, K.-H.; Ahn, H.-Y.; Lee, K.-D. Development of field scale model for estimating barley growth based on UAV NDVI and meteorological factors. SPIE 2018, 10783, 107831P. [Google Scholar]
- Karnieli, A.; Agam, N.; Pinker, R.T.; Anderson, M.; Imhoff, M.L.; Gutman, G.G.; Panov, N.; Goldberg, A. Use of NDVI and land surface temperature for drought assessment: Merits and limitations. J. Climate 2010, 23, 618–633. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using UAV based remote sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Paterson, A.H. High throughput phenotyping of cotton plant height using depth images under field conditions. Comput. Electron. Agric. 2016, 130, 57–68. [Google Scholar] [CrossRef]
- Rueda-Ayala, P.V.; Peña, M.J.; Höglind, M.; Bengochea-Guevara, M.J.; Andújar, D. Comparing UAV-based technologies and RGB-D reconstruction methods for plant height and biomass monitoring on grass ley. Sensors 2019, 19, 535. [Google Scholar] [CrossRef] [PubMed]
- Chao, Z.; Liu, N.; Zhang, P.; Ying, T.; Song, K. Estimation methods developing with remote sensing information for energy crop biomass: A comparative review. Biomass Bioenergy 2019, 122, 414–425. [Google Scholar] [CrossRef]
- Li, W.; Niu, Z.; Chen, H.; Li, D.; Wu, M.; Zhao, W. Remote estimation of canopy height and aboveground biomass of maize using high-resolution stereo images from a low-cost unmanned aerial vehicle system. Ecol. Indic. 2016, 67, 637–648. [Google Scholar] [CrossRef]









| Date | Season of the Year | Number of Plants Sampled | Data Acquisition | Sensors/Equipment |
|---|---|---|---|---|
| 9 May 2017 | Autumn | 475 | UAS flight, manual height and fresh and dry HY | Parrot Sequoia, ruler and manual cut |
| 4 July 2017 | Winter | |||
| 11 September 2017 | Early spring | |||
| 20 November 2017 | Late spring | |||
| 19 June 2018 | Winter | 426 | UAS flight, manual height, fresh and dry HY and ultrasonic sonar height | Parrot Sequoia, ruler, manual cut and ultrasonic sonar |
| 20 August 2018 | Late winter * | |||
| 23 October 2018 | spring | |||
| 20 November 2018 | Late spring |
| Date | Season | Overlap (Forward/Side) | Flight (m/s) | Flight Time (Minutes) | Georeferencing RMSE (m) | GSD (m/pixel) |
|---|---|---|---|---|---|---|
| 2017 | Autumn | 80%/75% | 6 | 4 | 0.02 | 0.02 |
| Winter | 80%/75% | 6 | 4 | 0.01 | 0.02 | |
| Early spring | 80%/75% | 6 | 4 | 0.01 | 0.02 | |
| Late spring | 80%/75% | 6 | 4 | 0.01 | 0.022 | |
| 2018 | Early winter | 75%/75% | 6 | 4 | 0.01 | 0.02 |
| Late winter | 75%/75% | 6 | 4 | 0.03 | 0.02 | |
| Early spring | 75%/75% | 6 | 4 | 0.04 | 0.02 | |
| Late spring | 75%/75% | 6 | 4 | 0.03 | 0.02 |
| 2017 | 2018 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CV Method | Partition | R2 | RMSE (g) | MAE | MPE% | R2 | RMSE (g) | MAE | MPE% |
| 2-folds | 2 | 0.63 | 32.81 | 24.31 | 22 | 0.69 | 23.80 | 15.21 | 30 |
| 5-folds | 5 | 0.64 | 32.66 | 24.30 | 22 | 0.69 | 23.67 | 15.11 | 30.63 |
| 10-folds | 10 | 0.64 | 32.60 | 24.29 | 22 | 0.69 | 23.71 | 15.16 | 30 |
| 20-folds | 20 | 0.64 | 32.59 | 24.30 | 22 | 0.70 | 23.50 | 15.15 | 30 |
| Random split * | 60%/40% | 0.64 | 32.77 | 24.34 | 22 | 0.69 | 23.96 | 15.50 | 30.70 |
| Random split * | 70%/30% | 0.64 | 32.77 | 24.33 | 22 | 0.69 | 23.92 | 15.49 | 30.68 |
| Random split * | 80%/20% | 0.64 | 32.71 | 24.31 | 22 | 0.69 | 23.91 | 15.49 | 30.66 |
| 2017 | 2018 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CV Method | Partition | R2 | RMSE (g) | MAE | MPE% | R2 | RMSE (g) | MAE | MPE% |
| 2-folds | 2 | 0.66 | 7.57 | 5.43 | 22 | 0.67 | 5.50 | 3.60 | 27.76 |
| 5-folds | 5 | 0.67 | 7.56 | 5.44 | 22 | 0.67 | 5.46 | 3.60 | 27.62 |
| 10-folds | 10 | 0.67 | 7.57 | 5.43 | 22 | 0.67 | 5.50 | 3.59 | 28 |
| 20-folds | 20 | 0.67 | 7.53 | 5.43 | 22 | 0.68 | 5.43 | 3.59 | 27.65 |
| Random split * | 60%/40% | 0.67 | 7.59 | 5.44 | 22 | 0.67 | 5.52 | 3.67 | 28.34 |
| Random split * | 70%/30% | 0.67 | 7.60 | 5.44 | 22 | 0.67 | 5.52 | 3.66 | 28.31 |
| Random split * | 80%/20% | 0.67 | 7.58 | 5.44 | 22 | 0.67 | 5.51 | 3.66 | 28.29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gebremedhin, A.; Badenhorst, P.; Wang, J.; Giri, K.; Spangenberg, G.; Smith, K. Development and Validation of a Model to Combine NDVI and Plant Height for High-Throughput Phenotyping of Herbage Yield in a Perennial Ryegrass Breeding Program. Remote Sens. 2019, 11, 2494. https://doi.org/10.3390/rs11212494
Gebremedhin A, Badenhorst P, Wang J, Giri K, Spangenberg G, Smith K. Development and Validation of a Model to Combine NDVI and Plant Height for High-Throughput Phenotyping of Herbage Yield in a Perennial Ryegrass Breeding Program. Remote Sensing. 2019; 11(21):2494. https://doi.org/10.3390/rs11212494
Chicago/Turabian StyleGebremedhin, Alem, Pieter Badenhorst, Junping Wang, Khageswor Giri, German Spangenberg, and Kevin Smith. 2019. "Development and Validation of a Model to Combine NDVI and Plant Height for High-Throughput Phenotyping of Herbage Yield in a Perennial Ryegrass Breeding Program" Remote Sensing 11, no. 21: 2494. https://doi.org/10.3390/rs11212494
APA StyleGebremedhin, A., Badenhorst, P., Wang, J., Giri, K., Spangenberg, G., & Smith, K. (2019). Development and Validation of a Model to Combine NDVI and Plant Height for High-Throughput Phenotyping of Herbage Yield in a Perennial Ryegrass Breeding Program. Remote Sensing, 11(21), 2494. https://doi.org/10.3390/rs11212494