Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification
Abstract
:1. Introduction
2. Proposed Methodology
2.1. Study Images
2.2. Frequency Transformation—Minimum Noise Fraction (MNF)
2.3. Frequency Transformation—Hilbert–Huang Transform (HHT)
2.4. Machine Learning Classification—Artificial Neural Networks (ANNs)
3. Results & Discussion
3.1. Frequency Transformation—MNF+HHT Transform
3.2. Machine Learning Classification—Training Sample Proportions
3.3. Machine Learning Classification—Neuron Numbers
4. Conclusions
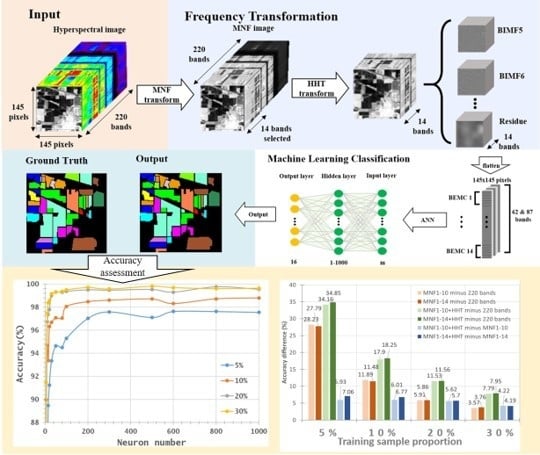
- With the aim of solving two critical issues in HSI classification, the curse of dimensionality and the limited availability of training samples, this study proposes a novel approach by integrating MNF and HHT transformations into ANN classification. MNF was performed to reduce the dimensionality of HSI, and the decomposition function of HHT produced more discriminative information from images. After MNF and HHT transformations, training samples were selected for each land cover type with four proportions and tested using 1–1000 neurons in an ANN. For a comparison purpose, three categories of image sets, the original HSI dataset, MNF-transformed images (two sets), and MNF+HHT-transformed images (two sets) were compared regarding their ANN classification performances.
- Two HSI datasets, the Indian Pines (IP) and Pavia University (PaviaU) datasets, were tested with the proposed method. The results showed that the IP MNF1–14+HHT-transformed images achieved the highest accuracy of 99.81% with a 30% training sample using 500 neurons, whereas the PaviaU dataset achieved the highest accuracy of 98.70% with a 30% training sample using 800 neurons. The results revealed that the proposed approach of integrating MNF and HHT transformations efficiently and significantly enhanced HSI classification performance by the ANN.
- In general, the classification accuracy increased as the training sample proportion increased and as the number of neurons increased, indicating the data-eager characteristics of ANNs. The MNF+HHT transformed image sets also displayed the highest accuracy statistically. A large accuracy improvement, 34.85%, was observed for the IP MNF1–14+HHT image set compared with the original 220 band IP image using 5% training samples. However, no significant difference was found between 20% and 30% training sample proportions, which demonstrates the limitations in the accuracy improvement that can be achieved by increasing the sample size. The accuracy improvement of the PaviaU dataset was smaller but still positive. For the PaviaU dataset, 10 MNFs showed superior performance to 14 MNFs when using 5% and 10% training samples, which reflected that 14 MNFs might include ineffective spectral information and thus decrease the classification accuracy. The PaviaU image set needed fewer MNFs than the IP set did to achieve a similar classification accuracy, due to its lower-dimensional spectral information
- Additionally, the accuracy improvement curve became relatively flat when more than 200 neurons were used for both datasets. This observation revealed that using more discriminative information from transformed images can reduce the number of neurons needed to adequately describe the data, as well as reducing the complexity of the ANN model.
- The proposed approach suggests new avenues for further research on HSI classification using ANNs. Various DL-based methods such as semantic segmentation [54], manifolding learning, GANs, RNN, SAE, SLFN, ELM, or automatic feature-extraction techniques could be further investigated as future possible research directions.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, M.-D.; Yang, Y.F.; Hsu, S.C. Application of remotely sensed data to the assessment of terrain factors affecting the Tsao-Ling landslide. Can. J. Remote Sens. 2004, 30, 593–603. [Google Scholar] [CrossRef]
- Yang, M.-D.; Su, T.C.; Hsu, C.H.; Chang, K.C.; Wu, A.M. Mapping of the 26 December 2004 tsunami disaster by using FORMOSAT-2 images. Int. J. Remote Sens. 2007, 28, 3071–3091. [Google Scholar] [CrossRef]
- Tsai, H.P.; Lin, Y.-H.; Yang, M.-D. Exploring Long Term Spatial Vegetation Trends in Taiwan from AVHRR NDVI3g Dataset Using RDA and HCA Analyses. Remote Sens. 2016, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Demir, B.; Erturk, S.; Güllü, M.K. Hyperspectral Image Classification Using Denoising of Intrinsic Mode Functions. IEEE Geosci. Remote Sens. Lett. 2010, 8, 220–224. [Google Scholar] [CrossRef]
- Taskin, G.; Kaya, H.; Bruzzone, L.; Kaya, G.T. Feature Selection Based on High Dimensional Model Representation for Hyperspectral Images. IEEE Trans. Image Process. 2017, 26, 2918–2928. [Google Scholar] [CrossRef]
- Romero, A.; Gatta, C.; Camps-Valls, G. Unsupervised Deep Feature Extraction for Remote Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2015, 54, 1349–1362. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Geng, J.; Wang, H. Hyperspectral image classification via contextual deep learning. EURASIP J. Image Video Process. 2015, 2015, 1778. [Google Scholar] [CrossRef] [Green Version]
- Kavzoglu, T. Increasing the accuracy of neural network classification using refined training data. Environ. Model. Softw. 2009, 24, 850–858. [Google Scholar] [CrossRef]
- Ratle, F.; Camps-Valls, G.; Weston, J. Semisupervised Neural Networks for Efficient Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2271–2282. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Tatnall, A.R.L. Introduction Neural networks in remote sensing. Int. J. Remote Sens. 1997, 18, 699–709. [Google Scholar] [CrossRef]
- Bruzzone, L.; Prieto, D. A technique for the selection of kernel-function parameters in RBF neural networks for classification of remote-sensing images. IEEE Trans. Geosci. Remote Sens. 1999, 37, 1179–1184. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep Learning-Based Classification of Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Guo, A.J.X.; Zhu, F. Spectral-Spatial Feature Extraction and Classification by ANN Supervised With Center Loss in Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2018, 57, 1755–1767. [Google Scholar] [CrossRef]
- Ahmad, M. A Fast 3D CNN for Hyperspectral Image Classification. arXiv 2020, arXiv:2004.14152. [Google Scholar]
- Zhang, H.; Li, Y.; Zhang, Y.; Shen, Q. Spectral-spatial classification of hyperspectral imagery using a dual-channel convolutional neural network. Remote Sens. Lett. 2017, 8, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Pantazi, X.E.; Moshou, D.; Bochtis, D. Intelligent Data Mining and Fusion Systems in Agriculture; Academic Press: Landon, UK, 2019. [Google Scholar]
- Ahmad, M.; Shabbir, S.; Oliva, D.; Mazzara, M.; Distefano, S. Spatial-prior generalized fuzziness extreme learning machine autoencoder-based active learning for hyperspectral image classification. Optik 2020, 206, 163712. [Google Scholar] [CrossRef]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep Convolutional Neural Networks for Hyperspectral Image Classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef] [Green Version]
- Ghamisi, P.; Chen, Y.; Zhu, X.X. A Self-Improving Convolution Neural Network for the Classification of Hyperspectral Data. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1537–1541. [Google Scholar] [CrossRef] [Green Version]
- Guidici, D.; Clark, M.L. One-Dimensional Convolutional Neural Network Land-Cover Classification of Multi-Seasonal Hyperspectral Imagery in the San Francisco Bay Area, California. Remote Sens. 2017, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Cui, Z.; Gan, Z.; Liu, F. Residual Group Channel and Space Attention Network for Hyperspectral Image Classification. Remote Sens. 2020, 12, 2035. [Google Scholar] [CrossRef]
- Lee, H.; Kwon, H. Contextual deep CNN based hyperspectral classification. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3322–3325. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–Spatial Classification of Hyperspectral Imagery with 3D Convolutional Neural Network. Remot. Sens. 2017, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Haut, J.M.; Paoletti, M.E.; Plaza, J.; Li, J.; Plaza, J. Active Learning With Convolutional Neural Networks for Hyperspectral Image Classification Using a New Bayesian Approach. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6440–6461. [Google Scholar] [CrossRef]
- Petersson, H.; Gustafsson, D.; Bergstrom, D. Hyperspectral image analysis using deep learning—A review. In Proceedings of the 2016 6th International Conference on Image Processing Theory, Tools and Applications (IPTA), Oulu, Finland, 12–15 December 2016. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Bhateja, V.; Tripathi, A.; Gupta, A. An Improved Local Statistics Filter for Denoising of SAR Images. In Recent Advances in Intelligent Informatics; Springer: Heidelberg, Germany, 2014; pp. 23–29. [Google Scholar] [CrossRef]
- Ahmad, M. Fuzziness-based Spatial-Spectral Class Discriminant Information Preserving Active Learning for Hyperspectral Image Classification. arXiv 2020, arXiv:2005.14236. [Google Scholar]
- Rasti, B.; Hong, D.; Hang, R.; Ghamisi, P.; Kang, X.; Chanussot, J.; Benediktsson, J.A. Feature Extraction for Hyperspectral Imagery: The Evolution from Shallow to Deep (Overview and Toolbox). IEEE Geosci. Remote Sens. Mag. 2020. [Google Scholar] [CrossRef]
- Hong, D.; Wu, X.; Ghamisi, P.; Chanussot, J.; Yokoya, N.; Zhu, X.X. Invariant Attribute Profiles: A Spatial-Frequency Joint Feature Extractor for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3791–3808. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhao, B.; Jia, X.; Liao, W.; Zhang, B. Optimized Kernel Minimum Noise Fraction Transformation for Hyperspectral Image Classification. Remote Sens. 2017, 9, 548. [Google Scholar] [CrossRef] [Green Version]
- Brémaud, P. Fourier Transforms of Stable Signals. In Mathematical Principles of Signal Processing: Fourier and Wavelet Analysis; Springer Science & Business Media: New York, NY, USA, 2013; pp. 7–16. [Google Scholar] [CrossRef]
- Yang, M.-D.; Su, T.-C.; Pan, N.-F.; Liu, P. Feature extraction of sewer pipe defects using wavelet transform and co-occurrence matrix. Int. J. Wavelets Multiresolut. Inf. Process. 2011, 9, 211–225. [Google Scholar] [CrossRef]
- Xia, J.; Du, P.; He, X.; Chanussot, J. Hyperspectral Remote Sensing Image Classification Based on Rotation Forest. IEEE Geosci. Remote Sens. Lett. 2014, 11, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.-D.; Su, T.-C. Segmenting ideal morphologies of sewer pipe defects on CCTV images for automated diagnosis. Expert Syst. Appl. 2009, 36, 3562–3573. [Google Scholar] [CrossRef]
- Su, T.-C.; Yang, M.-D.; Wu, T.-C.; Lin, J.-Y. Morphological segmentation based on edge detection for sewer pipe defects on CCTV images. Expert Syst. Appl. 2011, 38, 13094–13114. [Google Scholar] [CrossRef]
- Fauvel, M.; Benediktsson, J.A.; Chanussot, J.; Sveinsson, J.R. Spectral and Spatial Classification of Hyperspectral Data Using SVMs and Morphological Profiles. IEEE Trans. Geosci. Remote Sens. 2008, 46, 3804–3814. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Chen, G.; Tian, L.; Qin, K.; Qian, S.-E. Minimum Noise Fraction versus Principal Component Analysis as a Preprocessing Step for Hyperspectral Imagery Denoising. Can. J. Remote Sens. 2016, 42, 106–116. [Google Scholar] [CrossRef]
- Sun, Y.; Fu, Z.; Fan, L. A Novel Hyperspectral Image Classification Pattern Using Random Patches Convolution and Local Covariance. Remote Sens. 2019, 11, 1954. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.E.; Shen, Z.; Long, S.R.; Wu, M.C.; Shih, H.H.; Zheng, Q.; Yen, N.-C.; Tung, C.C.; Liu, H.H. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. A Math. Phys. Eng. Sci. 1998, 454, 903–995. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, G.; Gan, S.; Zhang, C. Enhancing seismic reflections using empirical mode decomposition in the flattened domain. J. Appl. Geophys. 2015, 119, 99–105. [Google Scholar] [CrossRef]
- Chen, Y.K.; Zhou, C.; Yuan, J.; Jin, Z.Y. Applications of empirical mode decomposition in random noise attenuation of seismic data. J. Seism. Explor. 2014, 23, 481–495. [Google Scholar]
- Chen, Y.; Ma, J. Random noise attenuation by f-x empirical-mode decomposition predictive filtering. Geophysics 2014, 79, V81–V91. [Google Scholar] [CrossRef]
- Linderhed, A. 2D empirical mode decompositions in the spirit of image compression. Wavelet Indep. Compon. Anal. Appl. IX 2002, 4738, 1–9. [Google Scholar] [CrossRef]
- Bhuiyan, S.M.A.; Adhami, R.R.; Khan, J. Fast and Adaptive Bidimensional Empirical Mode Decomposition Using Order-Statistics Filter Based Envelope Estimation. EURASIP J. Adv. Signal Process. 2008, 2008, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.-D.; Huang, K.-S.; Yang, Y.F.; Lu, L.-Y.; Feng, Z.-Y.; Tsai, H.P. Hyperspectral Image Classification Using Fast and Adaptive Bidimensional Empirical Mode Decomposition With Minimum Noise Fraction. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1950–1954. [Google Scholar] [CrossRef]
- Yang, M.-D.; Su, T.-C.; Pan, N.-F.; Yang, Y.-F. Systematic image quality assessment for sewer inspection. Expert Syst. Appl. 2011, 38, 1766–1776. [Google Scholar] [CrossRef]
- Bernabé, S.; Marpu, P.; Plaza, J.; Mura, M.D.; Benediktsson, J.A. Spectral–Spatial Classification of Multispectral Images Using Kernel Feature Space Representation. IEEE Geosci. Remote Sens. Lett. 2014, 11, 288–292. [Google Scholar] [CrossRef]
- Nielsen, A.A. Kernel Maximum Autocorrelation Factor and Minimum Noise Fraction Transformations. IEEE Trans. Image Process. 2010, 20, 612–624. [Google Scholar] [CrossRef] [Green Version]
- Trusiak, M.; Wielgus, M.; Patorski, K. Advanced processing of optical fringe patterns by automated selective reconstruction and enhanced fast empirical mode decomposition. Opt. Lasers Eng. 2014, 52, 230–240. [Google Scholar] [CrossRef]
- Park, K.; Hong, Y.K.; Kim, G.H.; Lee, J. Classification of apple leaf conditions in hyper-spectral images for diagnosis of Marssonina blotch using mRMR and deep neural network. Comput. Electron. Agric. 2018, 148, 179–187. [Google Scholar] [CrossRef]
- Shang, Y.; Wah, B.W. Global optimization for neural network training. Computer 1996, 29, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.-D.; Tseng, H.-H.; Hsu, Y.-C.; Tsai, H.P. Semantic Segmentation Using Deep Learning with Vegetation Indices for Rice Lodging Identification in Multi-date UAV Visible Images. Remote Sens. 2020, 12, 633. [Google Scholar] [CrossRef] [Green Version]














| Image | Indian Pine MNF1–10+HHT Training Sample Proportions | ||||
|---|---|---|---|---|---|
| Neuron Numbers | 5% | 10% | 20% | 30% | |
| 1 | 23.85 | 23.85 | 23.85 | 47.45 | |
| 5 | 81.54 | 88.31 | 90.40 | 90.74 | |
| 10 | 89.49 | 94.27 | 95.41 | 97.14 | |
| 15 | 90.08 | 95.73 | 97.67 | 97.97 | |
| 20 | 90.08 | 95.65 | 98.30 | 99.06 | |
| 30 | 92.22 | 97.41 | 98.61 | 98.94 | |
| 50 | 95.17 | 96.10 | 98.36 | 98.84 | |
| 80 | 95.40 | 96.46 | 99.07 | 99.23 | |
| 100 | 95.94 | 96.24 | 99.37 | 99.31 | |
| 200 | 96.24 | 97.90 | 99.40 | 99.20 | |
| 300 | 96.27 | 98.40 | 99.19 | 99.56 | |
| 500 | 96.72 | 98.24 | 99.48 | 99.60 | |
| 600 | 96.24 | 98.41 | 99.61 | 99.57 | |
| 800 | 96.94 | 98.91 | 99.37 | 99.47 | |
| 1000 | 96.88 | 98.72 | 99.57 | 99.54 | |
| Paired T test | 5% vs. 10%: p-value 0.000235993 (α = 0.01) | ||||
| 10% vs. 20%: p-value 0.00000956567 (α = 0.01) | |||||
| Image | Indian Pine MNF1–14+HHT Training Sample Proportions | ||||
|---|---|---|---|---|---|
| Neuron Numbers | 5% | 10% | 20% | 30% | |
| 1 | 23.85 | 44.31 | 47.39 | 47.66 | |
| 5 | 78.82 | 89.99 | 83.75 | 91.49 | |
| 10 | 83.73 | 94.70 | 96.74 | 97.45 | |
| 15 | 89.48 | 93.36 | 97.38 | 98.36 | |
| 20 | 91.25 | 96.31 | 97.80 | 98.54 | |
| 30 | 93.35 | 96.69 | 99.01 | 99.15 | |
| 50 | 94.63 | 97.07 | 99.31 | 99.29 | |
| 80 | 94.51 | 97.04 | 99.28 | 99.36 | |
| 100 | 95.30 | 98.06 | 99.37 | 99.50 | |
| 200 | 97.02 | 98.47 | 99.50 | 99.70 | |
| 300 | 97.59 | 98.60 | 99.47 | 99.57 | |
| 500 | 97.11 | 98.70 | 99.53 | 99.81 | |
| 600 | 97.62 | 98.31 | 99.30 | 99.69 | |
| 800 | 97.62 | 98.72 | 99.76 | 99.60 | |
| 1000 | 97.55 | 98.80 | 99.55 | 99.64 | |
| Paired T test | 5% vs. 10%: p-value 0.00543679 (α = 0.01) | ||||
| 10% vs. 20%: p-value 0.0589095 (α = 0.10) | |||||
| Image | PaviaU MNF1–10+HHT Training Sample Proportions | ||||
|---|---|---|---|---|---|
| Neuron Number | 5% | 10% | 20% | 30% | |
| 1 | 43.60 | 65.53 | 59.00 | 43.60 | |
| 5 | 86.27 | 89.53 | 87.33 | 90.16 | |
| 10 | 93.50 | 94.60 | 94.97 | 95.44 | |
| 15 | 93.99 | 94.79 | 96.25 | 95.88 | |
| 20 | 93.65 | 96.28 | 96.24 | 96.28 | |
| 30 | 95.09 | 95.53 | 96.55 | 96.97 | |
| 50 | 93.23 | 96.36 | 97.40 | 97.46 | |
| 80 | 93.85 | 95.91 | 97.16 | 97.34 | |
| 100 | 93.85 | 96.09 | 97.02 | 97.17 | |
| 200 | 93.58 | 95.78 | 97.08 | 97.64 | |
| 300 | 93.49 | 95.64 | 97.02 | 97.66 | |
| 500 | 93.47 | 95.87 | 97.24 | 97.82 | |
| 600 | 93.76 | 95.89 | 97.61 | 97.85 | |
| 800 | 93.60 | 96.86 | 96.90 | 97.55 | |
| 1000 | 92.97 | 96.09 | 97.08 | 98.22 | |
| Paired T test | 5% vs. 10%: p-value 0.0193299 (α = 0.05) | ||||
| Image | PaviaU MNF1–14+HHT Training Sample Proportions | ||||
|---|---|---|---|---|---|
| Neuron Number | 5% | 10% | 20% | 30% | |
| 1 | 66.12 | 66.19 | 65.86 | 65.99 | |
| 5 | 90.39 | 87.66 | 92.19 | 90.43 | |
| 10 | 93.68 | 95.34 | 96.05 | 95.45 | |
| 15 | 92.53 | 95.48 | 96.93 | 96.69 | |
| 20 | 93.73 | 96.17 | 96.50 | 97.46 | |
| 30 | 92.46 | 96.43 | 97.07 | 97.56 | |
| 50 | 92.91 | 96.25 | 97.73 | 98.04 | |
| 80 | 93.00 | 95.62 | 97.64 | 98.14 | |
| 100 | 93.03 | 95.58 | 97.54 | 97.98 | |
| 200 | 92.44 | 95.58 | 97.36 | 97.97 | |
| 300 | 92.24 | 95.99 | 97.46 | 98.27 | |
| 500 | 92.28 | 95.02 | 97.56 | 98.68 | |
| 600 | 93.75 | 95.69 | 97.93 | 98.31 | |
| 800 | 93.58 | 95.67 | 97.62 | 98.70 | |
| 1000 | 93.71 | 95.95 | 97.47 | 98.07 | |
| Paired T test | 5% vs. 10%: p-value 0.000154062 (α = 0.001) | ||||
| 10% vs. 20%: p-value 0.0000633683 (α = 0.001) | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.-D.; Huang, K.-H.; Tsai, H.-P. Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification. Remote Sens. 2020, 12, 2327. https://doi.org/10.3390/rs12142327
Yang M-D, Huang K-H, Tsai H-P. Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification. Remote Sensing. 2020; 12(14):2327. https://doi.org/10.3390/rs12142327
Chicago/Turabian StyleYang, Ming-Der, Kai-Hsiang Huang, and Hui-Ping Tsai. 2020. "Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification" Remote Sensing 12, no. 14: 2327. https://doi.org/10.3390/rs12142327
APA StyleYang, M. -D., Huang, K. -H., & Tsai, H. -P. (2020). Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification. Remote Sensing, 12(14), 2327. https://doi.org/10.3390/rs12142327