Mapping Vegetation at Species Level with High-Resolution Multispectral and Lidar Data Over a Large Spatial Area: A Case Study with Kudzu
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Target Vegetation Species
2.2. Data Details
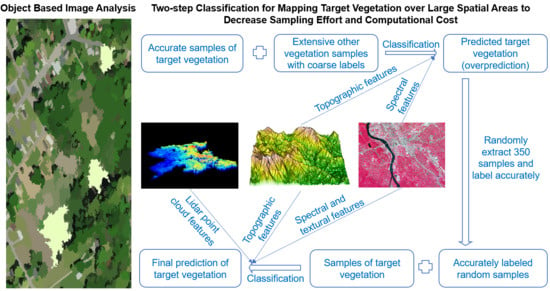
2.3. Designed Workflow
2.3.1. Image Segmentation
2.3.2. Vegetation Extraction and Coarsely Labeling Vegetation Objects
2.3.3. Integration of Light Detection and Ranging (Lidar) Point Clouds
2.3.4. Two-Step Classification
2.4. Comparison of the Proposed Method with the Traditional Method
2.5. Feature Selection, Accuracy Assessment, and Impact of Sample Size of Kudzu
3. Results
3.1. First-Step Classification
3.2. Second-Step Classification
3.3. Feature Importance
3.4. Impact of Kudzu Sample Size on Classification Accuracy
4. Discussion
4.1. Integration of Multiple Sources of Remote-Sensing Data for Vegetation Mapping
4.2. Decrease of Computational Cost and Sampling Effort
4.3. Sub-Sampling Improves Sample Specificity for Target Vegetation Species
4.4. Impact of Sample Size of Target Vegetation Species
4.5. Use and Limitation of the Proposed Workflow
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liang, W.; Tran, L.; Wiggins, G.J.; Grant, J.F.; Stewart, S.D.; Washington-Allen, R. Determining spread rate of kudzu bug (Hemiptera: Plataspidae) and its associations with environmental factors in a heterogeneous landscape. Environ. Entomol. 2019, 48, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Wardlow, B.; Egbert, S.; Kastens, J. Analysis of time-series MODIS 250 m vegetation index data for crop classification in the U.S. central great plains. Remote Sens. Environ. 2007, 108, 290–310. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, M.S.; Wilson, E.H.; Barrett, N.; Civco, D.L.; Prisloe, S.; Hurd, J.D.; Chadwick, C. Integrating multi-temporal spectral and structural information to map wetland vegetation in a lower Connecticut River tidal marsh. Remote Sens. Environ. 2008, 112, 4048–4060. [Google Scholar] [CrossRef]
- Yang, J.; Weisberg, P.J.; Bristow, N.A. Landsat remote sensing approaches for monitoring long-term tree cover dynamics in semi-arid woodlands: Comparison of vegetation indices and spectral mixture analysis. Remote Sens. Environ. 2012, 119, 62–71. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, X.; Xu, Y.; Wang, S.; Jia, T.; Hu, X.; Zhao, J.; Wei, L.; Zhang, L. Mini-UAV-borne hyperspectral remote sensing: From observation and processing to applications. IEEE Geosci. Remote Sens. Mag. 2018, 6, 46–62. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Tao, D.; Huang, X. On combining multiple features for hyperspectral remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2012, 50, 879–893. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of herbaceous vegetation using airborne hyperspectral imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Taboada, F.; Paredes, C.; Julián-Pelaz, J. Mapping of the invasive species Hakea sericea using unmanned aerial vehicle (UAV) and WorldView-2 imagery and an object-oriented approach. Remote Sens. 2017, 9, 913. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, O.S.; Shemrock, A.; Chabot, D.; Dillon, C.; Williams, G.; Wasson, R.; Franklin, S.E. Hierarchical land cover and vegetation classification using multispectral data acquired from an unmanned aerial vehicle. Int. J. Remote Sens. 2017, 38, 2037–2052. [Google Scholar] [CrossRef]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-based detailed vegetation classification with airborne high spatial resolution remote sensing imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef] [Green Version]
- Fu, B.; Wang, Y.; Campbell, A.; Li, Y.; Zhang, B.; Yin, S.; Xing, Z.; Jin, X. Comparison of object-based and pixel-based Random Forest algorithm for wetland vegetation mapping using high spatial resolution GF-1 and SAR data. Ecol. Indic. 2017, 73, 105–117. [Google Scholar] [CrossRef]
- Erker, T.; Wang, L.; Lorentz, L.; Stoltman, A.; Townsend, P.A. A statewide urban tree canopy mapping method. Remote Sens. Environ. 2019, 229, 148–158. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J.; Im, J. Synergistic use of QuickBird multispectral imagery and LIDAR data for object-based forest species classification. Remote Sens. Environ. 2010, 114, 1141–1154. [Google Scholar] [CrossRef]
- MacFaden, S.W.; O’Neil-Dunne, J.P.M.; Royar, A.R.; Lu, J.W.T.; Rundle, A.G. High-resolution tree canopy mapping for New York city using LIDAR and object-based image analysis. J. Appl. Remote Sens. 2012, 6, 063567. [Google Scholar] [CrossRef]
- Suárez, J.C.; Ontiveros, C.; Smith, S.; Snape, S. Use of airborne LiDAR and aerial photography in the estimation of individual tree heights in forestry. Comput. Geosci. 2005, 31, 253–262. [Google Scholar] [CrossRef]
- Dorigo, W.; Lucieer, A.; Podobnikar, T.; Čarni, A. Mapping invasive Fallopia japonica by combined spectral, spatial, and temporal analysis of digital orthophotos. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 185–195. [Google Scholar] [CrossRef]
- Nguyen, U.; Glenn, E.P.; Dang, T.D.; Pham, L.T.H. Mapping vegetation types in semi-arid riparian regions using random forest and object-based image approach: A case study of the Colorado River ecosystem, grand canyon, Arizona. Ecol. Inform. 2019, 50, 43–50. [Google Scholar] [CrossRef]
- Narumalani, S.; Mishra, D.R.; Wilson, R.; Reece, P.; Kohler, A. Detecting and mapping four invasive species along the floodplain of North Platte River, Nebraska. Weed Technol. 2009, 23, 99–107. [Google Scholar] [CrossRef]
- Maggiori, E.; Tarabalka, Y.; Charpiat, G.; Alliez, P. Convolutional neural networks for large-scale remote-sensing image classification. IEEE Trans. Geosci. Remote Sens. 2016, 55, 645–657. [Google Scholar] [CrossRef] [Green Version]
- Langford, Z.; Kumar, J.; Hoffman, F.; Breen, A.; Iversen, C. Arctic vegetation mapping using unsupervised training datasets and convolutional neural networks. Remote Sens. 2019, 11, 69. [Google Scholar] [CrossRef] [Green Version]
- Ristin, M.; Gall, J.; Guillaumin, M.; Van Gool, L. From categories to subcategories: Large-scale image classification with partial class label refinement. In Proceedings of the IEEE International Conference on Computer Vision Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 231–239. [Google Scholar]
- Lei, J.; Guo, Z.; Wang, Y. Weakly supervised image classification with coarse and fine labels. In Proceedings of the 14th Conference on Computer Robot Vision, Edmonton, AB, Canada, 17–19 May 2017; pp. 240–247. [Google Scholar]
- Sun, J.-H.; Liu, Z.-D.; Britton, K.O.; Cai, P.; Orr, D.; Hough-Goldstein, J. Survey of phytophagous insects and foliar pathogens in China for a biocontrol perspective on kudzu, Pueraria montana var. lobata (Willd.) Maesen and S. Almeida (Fabaceae). Biol. Control 2006, 36, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Forseth, I.N.; Innis, A.F. Kudzu (Pueraria montana): History, physiology, and ecology combine to make a major ecosystem threat. Crit. Rev. Plant Sci. 2004, 23, 401–413. [Google Scholar] [CrossRef]
- Follak, S. Potential distribution and environmental threat of Pueraria lobata. Open Life Sci. 2011, 6, 457–469. [Google Scholar] [CrossRef]
- Cheng, Y.-B.; Tom, E.; Ustin, S.L. Mapping an invasive species, kudzu (Pueraria montana), using hyperspectral imagery in western Georgia. J. Appl. Remote Sens. 2007, 1, 013514. [Google Scholar] [CrossRef]
- Li, J.; Bruce, L.M.; Byrd, J.; Barnett, J. Automated detection of Pueraria montana (kudzu) through Haar analysis of hyperspectral reflectance data. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Sydney, Australia, 9–13 July 2001; Volume 5, pp. 2247–2249. [Google Scholar]
- Colditz, R.R. An evaluation of different training sample allocation schemes for discrete and continuous land cover classification using decision tree-based algorithms. Remote Sens. 2015, 7, 9655–9681. [Google Scholar] [CrossRef] [Green Version]
- Heydari, S.S.; Mountrakis, G. Effect of classifier selection, reference sample size, reference class distribution and scene heterogeneity in per-pixel classification accuracy using 26 Landsat sites. Remote Sens. Environ. 2018, 204, 648–658. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M. On the importance of training data sample selection in random forest image classification: A case study in peatland ecosystem mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef] [Green Version]
- Ball, J.E.; Anderson, D.T.; Chan, C.S. Comprehensive survey of deep learning in remote sensing: Theories, tools, and challenges for the community. J. Appl. Remote Sens. 2017, 11, 042609. [Google Scholar] [CrossRef] [Green Version]
- ESRI. ArcGIS Desktop and Spatial Analyst Extension: Release 10.1; Environmental Systems Research Institute: Redlands, CA, USA, 2012. [Google Scholar]
- Oshiro, T.M.; Perez, P.S.; Baranauskas, J.A. How many trees in a random forest? In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2012; Volume 7376, pp. 154–168. [Google Scholar]
- Ramezan, A.C.; Warner, A.T.; Maxwell, E.A. Evaluation of sampling and cross-validation tuning strategies for regional-scale machine learning classification. Remote Sens. 2019, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Yeh, A.G.-O.; Li, X.; Lin, Z. A novel algorithm for land use and land cover classification using RADARSAT-2 polarimetric SAR data. Remote Sens. Environ. 2012, 118, 21–39. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, J.; Gong, J. UAV Remote sensing for urban vegetation mapping using random forest and texture analysis. Remote Sens. 2015, 7, 1074–1094. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Juel, A.; Groom, G.B.; Svenning, J.-C.; Ejrnæs, R. Spatial application of random forest models for fine-scale coastal vegetation classification using object based analysis of aerial orthophoto and DEM data. Int. J. Appl. Earth Obs. Geoinf. 2015, 42, 106–114. [Google Scholar] [CrossRef]
- Colditz, R.R.; Schmidt, M.; Conrad, C.; Hansen, M.C.; Dech, S. Land cover classification with coarse spatial resolution data to derive continuous and discrete maps for complex regions. Remote Sens. Environ. 2011, 115, 3264–3275. [Google Scholar] [CrossRef]
- Jin, H.; Stehman, S.V.; Mountrakis, G. Assessing the impact of training sample selection on accuracy of an urban classification: A case study in Denver, Colorado. Int. J. Remote Sens. 2014, 35, 2067–2081. [Google Scholar] [CrossRef]







| Data Source and Description | Feature Category | Extracted Features | Number of Features |
|---|---|---|---|
| National agricultural imagery program, Red-green-blue (RGB) and infrared spectral bands, 1 m resolution | Spectral bands | Mean value of 4-bands | 16 |
| Vegetation index | Mean, range, and standard deviation (SD) of normalized difference vegetation index (NDVI) | 12 | |
| Textural features on infrared band | Entropy, SD, and focal SD | 40 | |
| Gray-level co-occurrence matrix (GLCM) mean | |||
| GLCM variance | |||
| GLCM homogeneity | |||
| GLCM dissimilarity | |||
| GLCM contrast | |||
| GLCM entropy | |||
| GLCM second moment | |||
| 3D elevation program, Elevation, 2.5 m resolution | Topographic features | Mean and SD of elevation | 4 |
| Mean focal range and SD of elevation | |||
| 3D elevation program, Lidar point clouds, 3 returns/m2 | Canopy structural features | Coefficient of variation (CV), mean, variance, SD, intensity, and mode of Z-values of first return points | 8 |
| Mode of all return points | |||
| Density of all return points | |||
| Segments | Geometric feature | Area of each object | 1 |
| National land cover database, 30 m resolution | Coarse land cover label | Majority land-cover type of all pixels in each segment | NA |
| Class ID | Class Name | Percentage (%) | Class ID | Class Name | Percentage (%) |
|---|---|---|---|---|---|
| 11 | Open water | 0.33 | 42 | Evergreen forest | 2.67 |
| 21 | Developed, open space | 19.49 | 43 | Mixed forest | 9.16 |
| 22 | Developed, low intensity | 8.95 | 52 | Shrub/Scrub | 0.23 |
| 23 | Developed, medium intensity | 1.48 | 71 | Grassland/Herbaceous | 0.83 |
| 24 | Developed, high intensity | 0.08 | 81 | Pasture/Hay | 9.80 |
| 31 | Barren land | 0.03 | 82 | Cultivated crops | 0.01 |
| 41 | Deciduous forest | 46.69 | 90 | Woody wetlands | 0.23 |
| Number of Objects | Initial Objects | Preprocessing | Number of Predicted Kudzu | |||||
| Vegetation Extraction | Area Filtering | Proposed Method | Traditional Method | |||||
| 9,454,240 | 6,911,589 | 3,417,188 | 19,548 | 5,306 | 2,815 | 30,268 | ||
| OR | Testing data | 0% | 0% | 0% | 1% | 3% | 5% | 1% |
| Validation data | 0% | 0% | 0% | 1% | 3% | 5% | 1% | |
| Class | Proposed Method (First-Step Omission Rate) | Traditional Method | ||
|---|---|---|---|---|
| 1% | 3% | 5% | Second-Step Sampling | |
| Bare ground | 6 | 20 | 16 | 2 |
| Forest | 127 | 73 | 97 | 96 |
| Grass | 91 | 43 | 38 | 155 |
| Other herbaceous vegetation | 71 | 98 | 79 | 64 |
| Kudzu | 33 | 97 | 106 | 13 |
| Urban objects | 22 | 19 | 14 | 20 |
| Total number | 350 | 350 | 350 | 350 |
| Model | Proposed Method (First-Step Omission Rate) | Traditional Method | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1% (33 *) (612 **, 609 ***) | 3% (97 *) (1084 **, 1010 ***) | 5% (106 *) (853 **, 777 ***) | One-step sampling (50 *) (12826 **, 8791 ***) | Two-step sampling (50 *) (1726 **, 1129 ***) | |||||||||||
| PA | UA | K | PA | UA | K | PA | UA | K | PA | UA | K | PA | UA | K | |
| RF | 0.66 | 1.00 | 0.66 | 0.94 | 0.94 | 0.88 | 0.86 | 0.95 | 0.81 | 0.93 | 0.63 | 0.38 | 0.83 | 0.94 | 0.80 |
| SVM | 0.70 | 1.00 | 0.70 | 0.94 | 0.96 | 0.90 | 0.86 | 0.97 | 0.84 | 0.96 | 0.72 | 0.59 | 0.78 | 0.95 | 0.74 |
| Method | Model | Metrics | 50 | 100 | 150 | 200 |
|---|---|---|---|---|---|---|
| Proposed method with 1% OR at first-step classification | RF | PA | 0.79 | 0.96 | 0.96 | 0.96 |
| UA | 0.72 | 0.50 | 0.42 | 0.20 | ||
| SVM | PA | 0.84 | 0.93 | 0.96 | 0.96 | |
| UA | 0.84 | 0.82 | 0.60 | 0.56 | ||
| Traditional method with two-step sampling | RF | PA | 0.83 | 0.94 | 0.96 | 0.96 |
| UA | 0.50 | 0.40 | 0.40 | 0.24 | ||
| SVM | PA | 0.78 | 0.91 | 0.96 | 0.96 | |
| UA | 0.65 | 0.34 | 0.40 | 0.36 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, W.; Abidi, M.; Carrasco, L.; McNelis, J.; Tran, L.; Li, Y.; Grant, J. Mapping Vegetation at Species Level with High-Resolution Multispectral and Lidar Data Over a Large Spatial Area: A Case Study with Kudzu. Remote Sens. 2020, 12, 609. https://doi.org/10.3390/rs12040609
Liang W, Abidi M, Carrasco L, McNelis J, Tran L, Li Y, Grant J. Mapping Vegetation at Species Level with High-Resolution Multispectral and Lidar Data Over a Large Spatial Area: A Case Study with Kudzu. Remote Sensing. 2020; 12(4):609. https://doi.org/10.3390/rs12040609
Chicago/Turabian StyleLiang, Wanwan, Mongi Abidi, Luis Carrasco, Jack McNelis, Liem Tran, Yingkui Li, and Jerome Grant. 2020. "Mapping Vegetation at Species Level with High-Resolution Multispectral and Lidar Data Over a Large Spatial Area: A Case Study with Kudzu" Remote Sensing 12, no. 4: 609. https://doi.org/10.3390/rs12040609
APA StyleLiang, W., Abidi, M., Carrasco, L., McNelis, J., Tran, L., Li, Y., & Grant, J. (2020). Mapping Vegetation at Species Level with High-Resolution Multispectral and Lidar Data Over a Large Spatial Area: A Case Study with Kudzu. Remote Sensing, 12(4), 609. https://doi.org/10.3390/rs12040609