Machine Learning Algorithms to Predict Forage Nutritive Value of In Situ Perennial Ryegrass Plants Using Hyperspectral Canopy Reflectance Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
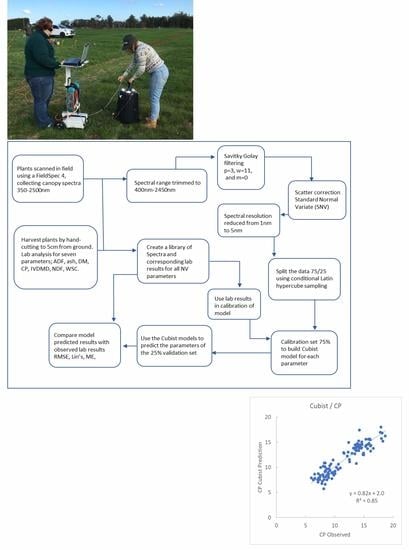
2.2. Spectra Collection
2.3. Spectra Data Pre-Processing
2.4. Splitting Data as Model Calibration and Validation
2.5. Spectral Model Development
2.6. Model Validation
2.7. Model Prediction of Nutritive Value (NV)
2.8. Model Variable Usage and Importance
2.9. Cubist Model Comparison to Partial Least Square Regression (PLSR) Model
3. Results
3.1. Descriptive Statistics and Evaluation of Model Performances for Key Nutritive Traits
3.2. Application of Models for High-Throughput NV Prediction
3.3. Key Model Drivers for Prediction
3.4. Cubist Model Comparison to PLSR Model
4. Discussion
4.1. Data Mining Techniques to Extract Biophysical Parameters of Perennial Ryegrass
4.2. Identify Specific Wavelengths Important for Modelling NV Parameters in Perennial Ryegrass
4.3. Evaluation of the Predictive Ability of Models Created Using Cubist to Analyze NV Parameters from an Independent Data Set
4.4. Advantages of the Data Mining Approach for NV Analysis as well as Potential Limiting Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blackburn, G.A. Hyperspectral Remote Sensing of Plant Pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [Green Version]
- Pullanagari, R.; Yule, I.; Hedley, M.; Tuohy, M.; Dynes, R.; King, W. Multi-spectral radiometry to Estimate Pasture Quality Components. Int. J. Adv. Precis. Agric. 2012, 13, 442–456. [Google Scholar] [CrossRef]
- Casler, M.D. Breeding Forage Crops for Increased Nutritional Value. Adv. Agron. 2001, 71, 51–107. [Google Scholar]
- Chapman, D.F.; Kenny, S.N.; Lane, N. Pasture and Forage Crop Systems for Non-irrigated Dairy Farms in Southern Australia: 3. Estimated Economic Value of Additional Home-grown Feed. Agric. Syst. 2011, 104, 589–599. [Google Scholar] [CrossRef]
- Smith, K.F.; Reed, K.F.M.; Foot, J.Z. An Assessment of the Relative Importance of Specific Traits for the Genetic Improvement of Nutritive Value in Dairy Pasture. Grass Forage Sci. 1997, 52, 167–175. [Google Scholar] [CrossRef]
- Mueller-Sim, T.; Jenkins, M.; Abel, J.; Kantor, G. The Robotanist: A Ground-based Agricultural Robot for High-throughput Crop Phenotyping. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA) (IEEE), Singapore, 29 May–3 June 2017; pp. 3634–3639. [Google Scholar]
- Casler, M.; Vogel, K. Accomplishments and Impact from Breeding for Increased Forage Nutritional Value. Crop Sci. 1999, 39, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Casler, M.D. Cultivar and Cultivar × Environment Effects on Relative Feed Value of Temperate Perennial Grasses. Crop Sci. 1990, 30, 722. [Google Scholar] [CrossRef]
- Richardson, A.D.; Reeves, J.B., III. Quantitative Reflectance Spectroscopy as an Alternative to Traditional Wet Lab Analysis of Foliar Chemistry: Near-infrared and Mid-infrared Calibrations Compared. Can. J. For. Res. 2005, 35, 1122–1130. [Google Scholar] [CrossRef]
- Starks, P.; Zhao, D.; Phillips, W.; Coleman, S. Development of Canopy Reflectance Algorithms for Real-Time Prediction of Bermudagrass Pasture Biomass and Nutritive Values. Crop Sci. 2006, 46, 927–934. [Google Scholar] [CrossRef] [Green Version]
- Araus, J.L.; Cairns, J.E. Field High-throughput Phenotyping: The New Crop Breeding Frontier. Trends Plant Sci. 2013, 19. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An Automated Robotic Field Phenotyping Platform for Detailed Crop Monitoring. Funct. Plant Biol. 2017, 44, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Zaman-Allah, M.; Vergara, O.; Araus, J.L.; Tarekegne, A.; Magorokosho, C.; Zarco-Tejada, P.J.; Hornero, A.; Albà, A.H.; Das, B.; Craufurd, P.; et al. Unmanned Aerial Platform-based Multi-spectral Imaging for Field Phenotyping of Maize. Plant Methods 2015, 11, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caicedo, J.P.R.; Verrelst, J.; Muñoz-Marí, J.; Moreno, J.; Camps-Valls, G. Toward a Semiautomatic Machine Learning Retrieval of Biophysical Parameters. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1249–1259. [Google Scholar] [CrossRef]
- Esteve Agelet, L.; Hurburgh, C.R. Limitations and Current Applications of Near Infrared Spectroscopy for Single Seed Analysis. Talanta 2014, 121, 288–299. [Google Scholar] [CrossRef]
- Li, Y.; Shao, X.; Cai, W. A Consensus Least Squares Support Vector Regression (LS-SVR) for Analysis of Near-infrared Spectra of Plant Samples. Talanta 2007, 72, 217–222. [Google Scholar] [CrossRef]
- Zhou, Z.; Morel, J.; Parsons, D.; Kucheryavskiy, S.V.; Gustavsson, A.-M. Estimation of Yield and Quality of Legume and Grass Mixtures Using Partial Least Squares and Support Vector Machine Analysis of Spectral Data. Comput. Electron. Agric. 2019, 162, 246–253. [Google Scholar] [CrossRef]
- Agelet, L.E.; Hurburgh, C.R. A Tutorial on Near Infrared Spectroscopy and Its Calibration. Crit. Rev. Anal. Chem. 2010, 40, 246–260. [Google Scholar] [CrossRef]
- Chen, H.; Pan, T.; Chen, J.; Lu, Q. Waveband Selection for NIR Spectroscopy Analysis of Soil Organic Matter Based on SG Smoothing and MWPLS Methods. Chemom. Intell. Lab. Syst. 2011, 107, 139–146. [Google Scholar] [CrossRef]
- Andueza, D.; Picard, F.; Jestin, M.; Andrieu, J.; Baumont, R. NIRS Prediction of the Feed Value of Temperate Forages: Efficacy of Four Calibration Strategies. Animal 2011, 5, 1002–1013. [Google Scholar] [CrossRef] [Green Version]
- Pullanagari, R.; Yule, I.; Tuohy, M.; Hedley, M.; Dynes, R.; King, W. In-field Hyperspectral Proximal Sensing for Estimating Quality Parameters of Mixed Pasture. Precis. Agric. 2012, 13, 351–369. [Google Scholar] [CrossRef]
- Smith, C.; Cogan, N.; Badenhorst, P.; Spangenberg, G.; Smith, K. Field Spectroscopy to Determine Nutritive Value Parameters of Individual Ryegrass Plants. Agronomy 2019, 9, 293. [Google Scholar] [CrossRef] [Green Version]
- Pasquini, C. Near infrared spectroscopy: A Mature Analytical Technique with New Perspectives—A Review. Anal. Chim. Acta 2018, 1026, 8–36. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.; Mouazen, A.M. Wheat Yield Prediction Using Machine Learning and Advanced Sensing techniques. Comput. Electron. Agric. 2016, 121, 57–65. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.-K.; Rumpf, T.; Römer, C.; Plümer, L. A Review of Advanced Machine Learning Methods for the Detection of Biotic Stress in Precision Crop Protection. An International J. Adv. Precis. Agric. 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Holmes, G.; Hall, M.; Prank, E. Generating Rule Sets from Model Trees. In Australasian Joint Conference on Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–12. [Google Scholar]
- Vapnik, V. Estimation of Dependences Based on Empirical Data; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Kuhn, M.; Weston, S.; Keefer, C.; Coulter, N. Cubist Models for Regression, R package Vignette R package version 0.0 2012, 18; CRAN: Vienna, Austria, 2012. [Google Scholar]
- Rossel, R.V.; Webster, R. Predicting Soil Properties from the Australian Soil Visible–near Infrared Spectroscopic Database. Eur. J. Soil Sci. 2012, 63, 848–860. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B.; Stockmann, U.; Hong, S.Y. Cubist, a Regression Rule Approach for use in Calibration of NIR Spectra. Picking Up Good Vib. 2013, 630. [Google Scholar]
- Padarian, J.; Minasny, B.; Mcbratney, A.B. Using Deep Learning for Digital Soil Mapping. SOIL 2019, 5, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Majeed, I.; Panigrahi, N.; Vasava, H.B.; Fidelis, C.; Karunaratne, S.; Bapiwai, P.; Yinil, D.; Sanderson, T.; Snoeck, D. Near Infrared Diffuse Reflectance Spectroscopy for Rapid and Comprehensive Soil Condition Assessment in Smallholder Cacao Farming Systems of Papua New Guinea. Catena 2019, 183, 104185. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Lin, L.I.K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Makdessi, N.A.; Jean, P.-A.; Ecarnot, M.; Gorretta, N.; Rabatel, G.; Roumet, P. How Plant Structure Impacts the Biochemical Leaf Traits Assessment from In-field Hyperspectral Images: A Simulation Study Based on Light Propagation Modeling in 3D Virtual Wheat Scenes. Field Crop. Res. 2017, 205, 95–105. [Google Scholar] [CrossRef]
- Doktor, D.; Lausch, A.; Spengler, D.; Thurner, M. Extraction of Plant Physiological Status from Hyperspectral Signatures Using Machine Learning Methods. Remote Sens. 2014, 6, 12247–12274. [Google Scholar] [CrossRef] [Green Version]
- Malmir, M.; Tahmasbian, I.; Xu, Z.; Farrar, M. Prediction of Macronutrients in Plant Leaves Using Chemometric Analysis and Wavelength Selection. J. Soils Sediments 2019, 1–11. [Google Scholar] [CrossRef]
- Biewer, S.; Fricke, T.; Wachendorf, M. Development of Canopy Reflectance Models to Predict Forage Quality of Legume-grass Mixtures. (Research) (Author abstract) (Report). Crop Sci. 2009, 49, 1917. [Google Scholar] [CrossRef]
- Thulin, S.M. Hyperspectral Remote Sensing of Temperate Pasture Quality. In Science, Engineering and Technology Portfolio; School of Mathematical and Geospatial Sciences, RMIT University Melbourne: Melbourne, Australia, 2008; p. 486. [Google Scholar]
- Wessman, C.A. Evaluation of Canopy Biochemistry. In Remote Sensing of Biosphere Functioning; Springer: Berlin/Heidelberg, Germany, 1990; pp. 135–156. [Google Scholar]
- Andueza, D.; Picard, F.; Martin-Rosset, W.; Aufrère, J. Near-infrared Spectroscopy Calibrations Performed on Oven-dried Green Forages for the Prediction of Chemical Composition and Nutritive Value of Preserved Forage for Ruminants. Appl. Spectrosc. 2016, 70, 1321–1327. [Google Scholar] [CrossRef]
- Danieli, P.P.; Carlini, P.; Bernabucci, U.; Ronchi, B. Quality Evaluation of Regional Forage Resources by Means of Near Infrared Reflectance Spectroscopy. Ital. J. Anim. Sci. 2004, 3, 363–376. [Google Scholar] [CrossRef]
- Zeng, L.; Chen, C. Using Remote Sensing to Estimate Forage Biomass and Nutrient Contents at Different Growth Stages. Biomass Bioenergy 2018, 115, 74–81. [Google Scholar] [CrossRef]
- Downey, G.; Robert, P.; Bertrand, D.; Devaux, M.F. Near Infra-red Analysis of Grass Silage by Principal Component Analysis of Transformed Reflectance Data. J. Sci. Food Agric. 1987, 41, 219–229. [Google Scholar] [CrossRef]
- Downey, G.; Robert, P.; Bertrand, D.; Devaux, M.F. Dried Grass Silage Analysis by NIR Reflectance Spectroscopy—A Comparison of Stepwise Multiple Linear and Principal Component Techniques for Calibration Development on Raw and Transformed Spectral Data. J. Chemom. 1989, 3, 397–407. [Google Scholar] [CrossRef]
- Ferner, J.; Linstädter, A.; Südekum, K.-H.; Schmidtlein, S. Spectral Indicators of Forage Quality in West Africa’s Tropical Savannas. Int. J. Appl. Earth Obs. Geoinf. 2015, 41, 99–106. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Q. Evaluation of Informative Bands Used in Different PLS Regressions for Estimating Leaf Biochemical Contents from Hyperspectral Reflectance. Remote Sens. 2019, 11, 197. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Lei, Y.; Louzada Prates, L.; Yu, P. Evaluation of Near-infrared (NIR) and Fourier transform mid-infrared (ATR-FT/MIR) Spectroscopy Techniques Combined with Chemometrics for the Determination of Crude Protein and Intestinal Protein Digestibility of Wheat. Food Chem. 2019, 272, 507–513. [Google Scholar] [CrossRef]
- Shorten, P.R.; Leath, S.R.; Schmidt, J.; Ghamkhar, K. Predicting the Quality of Ryegrass Using Hyperspectral Imaging. (Report). Plant Methods 2019, 15. [Google Scholar] [CrossRef] [Green Version]
- Balabin, R.M.; Safieva, R.Z.; Lomakina, E.I. Comparison of Linear and Nonlinear Calibration Models Based on Near Infrared (NIR) Spectroscopy Data for Gasoline Properties Prediction. Chemom. Intell. Lab. Syst. 2007, 88, 183–188. [Google Scholar] [CrossRef]
- Capolupo, A.; Kooistra, L.; Berendonk, C.; Boccia, L.; Suomalainen, J. Estimating Plant Traits of Grasslands from UAV-acquired Hyperspectral Images: A Comparison of Statistical Approaches. ISPRS Int. J. Geo Inf. 2015, 4, 2792–2820. [Google Scholar] [CrossRef]
- Chen, D.; Huang, J.; Jackson, T.J. Vegetation Water Content Estimation for Corn and Soybeans Using Spectral Indices Derived from MODIS Near- and Short-wave Infrared Bands. Remote Sens. Environ. 2005, 98, 225–236. [Google Scholar] [CrossRef]
- Da Silva, C.R.; Centeno, J.A.S.; Aranha, S.R. Reduction of the Dimensionality of Hyperspectral Data for the Classification of Agricultural Scenes. In Proceedings of the 13th Symposium Deformation Measurements and Analysis, Lisbon, Portugal, 12–15 May 2008. [Google Scholar]
- Shenk, J.S.; Workman, J.J., Jr.; Westerhaus, M.O. Application of NIR Spectroscopy to Agricultural Products. In Handbook of Near-Infrared Analysis; Burns, D.A., Ciurczak, E.W., Eds.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Tsenkova, R. Aquaphotomics: Dynamic spectroscopy of aqueous and biological systems describes peculiarities of water. J. Near Infrared Spectrosc. 2009, 17, 303–313. [Google Scholar] [CrossRef]
- Abrams, S.M.; Shenk, J.S.; Harpster, H.W. Potential of Near Infrared Reflectance Spectroscopy for Analysis of Silage Composition1,2,3. J. Dairy Sci. 1988, 71, 1955–1959. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of Variability in Canopy Reflectance and the Convergent Properties of Plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Tsenkova, R. Aquaphotomics: The Extended Water Mirror Effect Explains Why Small Concentrations of Protein in Solution can be Measured with Near Infrared Light. Nir News 2008, 19, 12–13. [Google Scholar] [CrossRef]
- Wijesingha, J.; Astor, T.; Schulze-Brüninghoff, D.; Wengert, M.; Wachendorf, M. Predicting Forage Quality of Grasslands Using UAV-Borne Imaging Spectroscopy. Remote Sens. 2020, 12, 126. [Google Scholar] [CrossRef] [Green Version]
- Goodchild, A.V.; El Haramein, F.J.; El Moneim, A.A.; Makkar, H.P.S.; Williams, P.C. Prediction of phenolics and tannins in forage legumes by near infrared reflectance. J. Near Infrared Spectrosc. 1998, 6, 7. [Google Scholar] [CrossRef]
- Mirik, M.; Norland, J.E.; Crabtree, R.L.; Biondini, M.E. Hyperspectral one-meter-resolution remote sensing in Yellowstone National Park, Wyoming: I. Forage nutritional values. Rangel. Ecol. Manag. 2005, 58, 452–458. [Google Scholar] [CrossRef]




| Parameter | R2 Calibration | R2 Validation | LCC Calibration | LCC Validation | MSE Calibration |
| ADF | 0.69 | 0.75 | 0.81 | 0.85 | 4.54 |
| Ash | 0.71 | 0.66 | 0.82 | 0.80 | 2.08 |
| IVDMD | 0.72 | 0.82 | 0.83 | 0.89 | 15.20 |
| NDF | 0.72 | 0.78 | 0.84 | 0.87 | 18.18 |
| CP | 0.82 | 0.74 | 0.89 | 0.85 | 2.73 |
| WSC | 0.60 | 0.49 | 0.73 | 0.68 | 6.20 |
| DM | 0.81 | 0.69 | 0.89 | 0.82 | 7.68 |
| Parameter | MSE Validation | RMSE Calibration | RMSE Validation | Bias Calibration | Bias Validation |
| ADF | 3.39 | 2.13 | 1.84 | 0.15 | -0.33 |
| Ash | 2.39 | 1.44 | 1.55 | -0.14 | -0.16 |
| IVDMD | 7.29 | 3.90 | 2.70 | 0.16 | 0.40 |
| NDF | 13.06 | 4.26 | 3.61 | 0.19 | -0.29 |
| CP | 4.08 | 1.65 | 2.02 | -0.04 | -0.13 |
| WSC | 7.68 | 2.49 | 2.77 | -0.06 | 0.27 |
| DM | 11.60 | 2.77 | 3.41 | 0.07 | 0.30 |
| ADF Calibration | ADF Prediction | Ash Calibration | Ash Prediction | IVDMD Calibration | IVDMD Prediction | NDF Calibration | |
| Average | 25.68 | 26.68 | 11.65 | 12.45 | 74.49 | 72.5 | 48.9 |
| Minimum | 17.64 | 14.76 | 5.69 | 6.76 | 47.31 | 40.09 | 33.73 |
| maximum | 41.37 | 46.72 | 23.47 | 21.79 | 83.41 | 87.97 | 76.90 |
| Standard Deviation | 3.62 | 4.30 | 2.61 | 2.16 | 6.38 | 7.87 | 7.71 |
| NDF Prediction | CP Calibration | CP Prediction | WSC Calibration | WSC Prediction | DM Calibration | DM Prediction | |
| Average | 49.17 | 14.05 | 14.92 | 22.07 | 21.12 | 26.13 | 28.18 |
| Minimum | 22.94 | 5.98 | 5.00 | 12.60 | 8.65 | 6.47 | 3.62 |
| maximum | 75.64 | 24.89 | 31.00 | 32.03 | 32.38 | 55.12 | 58.24 |
| Standard Deviation | 6.46 | 3.83 | 2.68 | 3.78 | 2.51 | 6.16 | 7.10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, C.; Karunaratne, S.; Badenhorst, P.; Cogan, N.; Spangenberg, G.; Smith, K. Machine Learning Algorithms to Predict Forage Nutritive Value of In Situ Perennial Ryegrass Plants Using Hyperspectral Canopy Reflectance Data. Remote Sens. 2020, 12, 928. https://doi.org/10.3390/rs12060928
Smith C, Karunaratne S, Badenhorst P, Cogan N, Spangenberg G, Smith K. Machine Learning Algorithms to Predict Forage Nutritive Value of In Situ Perennial Ryegrass Plants Using Hyperspectral Canopy Reflectance Data. Remote Sensing. 2020; 12(6):928. https://doi.org/10.3390/rs12060928
Chicago/Turabian StyleSmith, Chaya, Senani Karunaratne, Pieter Badenhorst, Noel Cogan, German Spangenberg, and Kevin Smith. 2020. "Machine Learning Algorithms to Predict Forage Nutritive Value of In Situ Perennial Ryegrass Plants Using Hyperspectral Canopy Reflectance Data" Remote Sensing 12, no. 6: 928. https://doi.org/10.3390/rs12060928
APA StyleSmith, C., Karunaratne, S., Badenhorst, P., Cogan, N., Spangenberg, G., & Smith, K. (2020). Machine Learning Algorithms to Predict Forage Nutritive Value of In Situ Perennial Ryegrass Plants Using Hyperspectral Canopy Reflectance Data. Remote Sensing, 12(6), 928. https://doi.org/10.3390/rs12060928