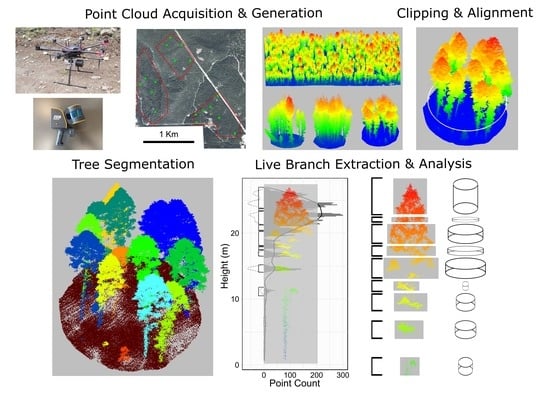
1. Introduction
While wildfires are critical disturbances that can help to maintain the health and diversity of a variety of ecosystems [
1], changes in global fire regimes are projected to have severe and long-lasting ecological repercussions. Depending on the particular geographic area, these changes include increases in fire frequency, size, and intensity and can be attributed to climate change, fire suppression, and changes in vegetation composition [
2]. Departures from historic fire regimes are projected to cause many forested ecosystems to have reductions in ecosystem resilience, a reduced capacity to provide ecosystem services, and to experience shifts in forest structure and composition [
3]. These changing fire regimes have already had substantial, multi-faceted socio-economic impacts, such as active suppression costs, damage to infrastructure, as well as short- and long-term impacts to human health [
4]. A recent literature review estimated the total economic burden of wildfire in the United States alone to be more than
$71.1 billion per year [
4].
To evaluate and mitigate the risks associated with wildfires, land managers make use of an array of wildfire behaviour models. These models predict potential fire behaviour using site-specific information about weather, topography, and fuels [
5]. Of the required fuel information, significant attention has been given to forest canopy fuels. Accurate measurements of canopy fuels are critical, as they are the primary fuel layer that supports the initiation and spread of crown fire [
6]. These measurements are typically derived from field-collected forest inventory data using allometric equations that produce stand-level fuel metrics [
5,
7]. The derivation of these critical metrics begins at the individual tree-level with an estimation of the amount and vertical arrangement of combustible fuels within individual tree crowns. This is typically calculated by deriving the total weight of crown foliage and fine fuels and distributing them from the base of the crown to the treetop [
5,
7].
While this approach is widely used in a variety of wildfire behaviour and fuel models, there are well-known limitations. First, the allometric equations assume that crown fuels are uniformly distributed within individual trees and do not typically incorporate any measure of crown width [
8]. This is an oversimplification that has been shown to impact fire behaviour [
9]. Additionally, due to the high cost of developing local allometries, generic equations are commonly applied, potentially resulting in unreliable predictions [
10,
11]. The second limitation revolves around the critical measure of live crown base height. This metric is used as the starting height to vertically distribute crown fuel, as well as to estimate the likelihood of fire propagating from the surface into tree crowns [
12]. Due to vague and differing definitions, this metric can be difficult to consistently measure in the field. Variation in this definition relates to how different authors define lower crown layers, the primary crown, and the vertical continuity between them. Early research did not always specify continuity and often defined live crown base height as the height of the lowest live branch [
13]. Others offer more detail and necessitate that these branches must provide a vertically continuous path into the primary crown [
14]. Even more detailed definitions followed, refining continuity by specifying that vertical gaps of up to ~1.5 m can exist within a “continuous” crown [
15].
Improving the estimation of crown fuels is an active area of research, although much of this research relies solely on field-based methods. While these studies are valuable, their applications are typically limited to relatively fine spatial scales. Alternatively, high density three-dimensional light detection and ranging (LiDAR) point clouds have been used to estimate tree- and stand-level fuel metrics. For over a decade, LiDAR point clouds acquired from conventional aircraft have been successfully used to estimate broad scale, stand-level canopy fuel metrics [
16,
17,
18]. Recently, more detailed and comprehensive characterizations of tree-level crown fuel have been undertaken using terrestrial or mobile laser scanning (TLS or MLS, respectively) [
19,
20,
21,
22]. The past five years has seen the acquisition of high-density LiDAR point clouds from remotely piloted aerial systems (RPAS). These types of point clouds can capture greater levels of detail than conventional aircraft-acquired LiDAR. These point clouds are not as dense as TLS or MLS point clouds but offer much greater spatial coverage. These characteristics result in RPAS LiDAR point clouds having the potential to characterize crown fuels at comparable detail to TLS but across broader spatial scales.
In this study, we examine the capability of high-density RPAS point clouds to characterize the vertical arrangement and volume of live crown fuels within individual trees. To accomplish this, we develop an automated method that detects and quantifies all branch structures that contain live foliage, hereafter “live branch clusters”. We apply our new method to over 100 individual trees with varying crown fuel loads from a dry, interior British Columbia forest system. Comparisons were made against manual measurements taken from MLS point clouds. The vertical arrangement of crown fuel is then summarized at the tree-level to describe crown fuel metrics in a comprehensive and systematic way. Using this approach, the amount and distribution of crown fuel can be accurately mapped across large numbers of trees, providing key fuel inputs for fire behaviour models.
3. Results
Below, we present the following results: cluster matching, cluster-level validation, and tree-level validation. The cluster matching results show the degree to which the automatically extracted and manually measured clusters aligned with one another. The cluster-level results show the level of agreement between the dimensions and vertical arrangements of the successfully matched clusters, while the tree-level results show the level of agreement between the extracted and measured accumulated metrics for each tree.
3.1. Cluster Matching
Table 1 summarizes statistics for all extracted and measured clusters, as well as the overall accuracy and commission and omission errors. In general, the automated method extracted many more clusters than the validation method. The extracted and measured clusters had similar mean diameters, but the extracted clusters tended to have much lower vertical length and volume values. The majority of automatically extracted clusters were successfully matched to a manually measured cluster (797 of 825 clusters). This resulted in a high overall accuracy (91.72%) with a low commission error (3.2%). Only 44 measured clusters did not correspond to an extracted cluster, resulting in an omission error of 5.06%. The lower mean heights of the FN clusters relative to all measured clusters indicate that these erroneous clusters tended to be located lower in the tree. The low mean and total volumes additionally indicate that these cluster were smaller in size than other manually measured clusters.
3.2. Cluster-Level Validation
The vertical arrangement of the extracted and measured clusters was in relatively close agreement (
Figure 6). The extracted upper boundaries corresponded well to the measured upper boundaries. This correspondence is exhibited by an R-squared value of 0.9926, low variance (0.6153 m), and a line of best fit that closely matches the 1:1 line. The extracted lower boundaries did not predict the measured values as well, with a lower R-squared value (0.6998) and higher variance (2.347 m). In many cases, the automated method substantially overpredicted the lower boundary of matched clusters, resulting in many points below the 1:1 line.
Figure 7 shows relatively strong agreement between the extracted and measured branch cluster dimensions. The extracted cluster diameters and volumes relate well to their manually measured counterparts, with respective R-squared values of 0.8725 and 0.9060. While still exhibiting a relatively strong relationship, the extracted vertical length of each cluster did not predict the measured values as well as the other two metrics. This deviation mirrors the relationship exhibited between the lower boundary of extracted and measured clusters shown in
Figure 6. In this instance, the automated method often underpredicts the vertical length of each cluster.
3.3. Tree-Level Validation
The focal point of
Figure 8 illustrates three pairs of MLS and RPAS LiDAR point clouds. Each pair represents a different tree (i, ii, iii) that was used within this study. The histograms, splines, second derivative lines, and extracted clusters shown here were calculated from the distribution of the RPAS LiDAR point height. Comparing these features with the MLS point clouds and the manually measured clusters highlights the level of detail provided by these two types of point clouds. The subpanels show relationships between the extracted and measured values for the tree-level metrics, highlighting the values for each of the three example trees.
The agreement between measured and extracted attributes were more variable at the tree-level than at the cluster-level. Most extracted metrics either under- or overpredict their respective validation metrics. The strongest match was observed for crown width, which had a line of best fit that closely matched the 1:1 line. The extracted and measured total live branch volumes corresponded strongly (R-squared of 0.9111), although a consistent underprediction can be observed. The remaining three metrics had moderate to weak relationships. Of these, the live crown length exhibited the strongest relationship. This metric had an R-squared value of 0.5568, although it tended to underpredict the measured values, especially for the smaller trees. Neither the height of the lowest live branch nor the live crown base height was accurately predicted using the automated method. For both these metrics, the automated method consistently assumed a higher height than was manually measured.
Among the three examples of the MLS and RPAS LiDAR point clouds, differences were greatest in lower portions of the crown, especially for tree iii. Tracing the extracted and measured values for tree iii across each of the five scatter plots shows how differences in lower crown point counts and densities impacted the results of this study. The automated method overpredicted the height of the lowest live branch and the live crown base height. This was primarily due to the relative absence of these lower crown points from the RPAS LiDAR point cloud. These overpredictions also corresponded to an underprediction for the vertical crown length. Tree ii had nearly all the same features present in both point clouds, but the automated method did not detect the lowest manually measured live branch. This resulted in a slight overprediction of the lowest live branch height. This cluster was present in the RPAS LiDAR point cloud, but not in great enough detail to be extracted by the automated method. Tree i also had nearly all the same branch clusters present in both point clouds, but the automated method was unable to detect the same gaps as the validation dataset. This resulted in a large underprediction of live crown base height, something that was not indicative of the general trend associated with live crown base height relationship.
3.4. Summary of Extracted Tree-Level Data
Figure 9 shows box and whisker plots for four tree-level metrics extracted from 104 trees using the automated method developed for this study. The values for live crown length had the largest range, followed by live crown base height and the height of the lowest live branch. Crown width values were most consistent, showing relatively little deviation from the median value of ~4.5 m. The overall and interquartile ranges of these four metrics were indicative of the variability observed in the sampled trees. This same variability was observed in the three example trees shown in
Figure 8. Each tree shown had variable amounts and arrangement of living fuel, while having relatively similar crown widths.
4. Discussion
This study evaluated the ability of RPAS LiDAR point clouds to characterize the vertical arrangement and volume of crown fuels within individual trees. The new method extracted and quantified branch structures that contained live fuel and compared cluster and tree-level measurements to manual measurements collected from MLS point clouds. The differences between the extracted and measured metrics revealed important strengths and weaknesses of both the method and the utility of both types of point clouds.
4.1. Cluster-Level Fuels
The cluster-level analysis supports the utility of RPAS LiDAR to characterize fine-scale, within-tree geometry at a level of detail similar to TLS or MLS point clouds. This is particularly true for the more dominant portions of the tree crowns. Over 96% of all automatically extracted live branch clusters were successfully matched to manually measured branch clusters, which represented a similarly large proportion of all extracted volume. While a smaller proportion of manually measured branch clusters were matched to extracted clusters (~79%), the unmatched clusters represented less than 2% of the measured volume.
Comparing the vertical arrangement and dimensions of matched clusters revealed strong correspondences between the two datasets. The vertical arrangement of each cluster can be described by the lower and upper boundaries, both of which were predicted well, albeit not as strongly for the lower boundaries. The overprediction of these lower boundaries was mirrored by a general underprediction of the measured vertical length of each cluster. The measured diameter of each cluster was also predicted quite well by the automated method and did so without an under or overprediction. The vertical length and diameter of each cluster were used to calculate a simple cylindrical volume for each cluster. Comparing the measured and extracted volumes revealed a strong correspondence. While these results only considered matched clusters, the favorable trends support the use of this method and data for the evaluation of within-tree geometry.
4.2. Tree-Level Fuels
The aggregation of cluster-level information at the tree-level revealed more nuanced information about how RPAS LiDAR and MLS point clouds compare. This aggregation also shows the degree to which this method can extract tree-level information. While the automated method presented here is not infallible, many of the shortcomings presented in the tree-level results can be attributed to inherent differences between MLS and RPAS LiDAR point clouds. The overall differences in the quality of each point cloud were evident at the onset of this study by the much higher density of MLS point clouds. There were additional differences related to where each point cloud was collected. Due to the ground-based system, the majority of the MLS points were located beneath the base of the live crown, whereas the majority of the RPAS LiDAR points were located above this level. These differences resulted in overpredictions of live crown base height and the height of the lowest live branches. These inaccuracies are also mirrored by an underprediction of crown length. The reduced level of detail in the lower crown can also be observed in nearly every point cloud shown above.
Of the tree-level metrics evaluated in this study, the strongest relationships were observed for crown width and total live branch volume. Both these values can be reliably extracted from dominant portions of the crown that tend to be readily visible from airborne platforms. While extracted total live branch volume exhibited a very strong relationship with the measured values, it did so with a strong overprediction. This overprediction is partially due to undetected live fuels in the lower crown, but more so due to instances where multiple extracted clusters were contained within a single measured cluster. The measured clusters would have only been represented by a single cylinder with a constant diameter. The extracted method would instead represent these same clusters with multiple smaller cylinders, only one of which would have the same diameter as the manually measured cluster. This difference in volumetric calculations resulted in large differences between the extracted and measured values.
Live crown base height was calculated in a way that is similar to existing field-based methods [
8]. While frequently applied, these methods can ignore fuel beneath the base of the primary crown, depending on the amount of vertical separation between fuel layers. In theory, accurate measures of live crown base height could be used in conjunction with crown width to calculate a simplistic crown volume. However, these types of methods would likely underrepresent the amount of fuel in lower portions of trees. Nearly 40% of the manually measured trees had living fuel between the ground and the base of the live crown. Of these trees, the average volume of fuel in this zone represented almost 12% of the total tree volume, and in some cases, over half the total volume.
This method was able to accurately characterize the locations and dimensions of the majority of live branch clusters, but not as reliably for lower clusters. This shortcoming is primarily a result of differences in the analyzed and reference data. Ground-based LiDAR is able to capture small features beneath the primary crown much more reliably than airborne LiDAR. Furthermore, the extremely low commission errors speak to the ability of the method to detect features that are present within a given point cloud. Therefore, the accurate extraction of these lower crown features is dependent on the quality and density of the provided point cloud. Overall, this work demonstrates a method that can reliably predict the amount and locations of large proportions of the total crown fuel present within a given tree.
4.3. Links to Past Research
The results of this study support and refine earlier work regarding the strengths and weaknesses of remotely derived airborne and ground-based point clouds. It has been shown that low density LiDAR point clouds collected from conventional aircraft can be used to successfully characterize stand-level overstory fuels [
16,
17,
18]. Although depending on point density and canopy cover, fuels below the primary crown are unable to be characterized to the same degree as ground-based point clouds [
38]. The results demonstrated here potentially extend this relationship to RPAS LiDAR point clouds, regardless of their high point density compared to conventional LiDAR point clouds. While not quantitatively evaluated in this study, tree segmented from RPAS LiDAR point clouds had more complete reconstructions in instances where the surrounding canopy cover was low. Similar observations were made for trees that had simpler crown structures.
The definition of crown base height that was used in this study allowed gaps of up to 25 cm to exist within a “continuous” crown. Similar methods as were presented here can be used to extract a flexible measurement of live crown base height with variable levels of gaps between live branches. This kind of measurement could represent a potentially significant outcome of this research. Crown base height definitions that rely on fixed gap thresholds assume that fire can only spread between crown layers separated by vertical gaps lower than the threshold. In this way, these definitions assume a fixed flame length, something that has long been understood to be variable depending on fuel and weather conditions [
39]. This research also supports earlier work [
9] by providing tools that can be used to model crown volume using non-uniform distributions of crown fuel. While contributing to this type of research, it is important to note that this work does not incorporate any information on the weights of available fuel, instead focusing on quantifying the three-dimensional volumes of portions of trees that contain available fuel.
4.4. Limitations and Future Work
The greatest limitation of this new method is the inability to extract lower crown fuels such as live crown base height and the height of the lowest live branches. While these missed branches were likely small in volume, their absence would likely result in an underprediction of true fire behaviour. This is especially true in cases where they are in close vertical proximity to other live crown fuels, thereby representing ladder fuels. Applying these methods directly to point clouds that have higher point densities beneath the primary crown would likely improve the characterization of these lower crown fuels. Typically, these types of point clouds are collected at the plot-level, thereby limiting their spatial scale. Recent advancements in RPAS and MLS systems have allowed the two to be combined for beneath canopy flights [
40], allowing MLS data to be collected across larger spatial scales.
The method developed in this study was only applied to trees that were well segmented, which were primarily composed of the more dominant, overstory trees. These segmentation results are supported by previous research, which has shown that perfectly segmenting non-dominant, subcanopy trees can be exceedingly difficult, if not impossible [
27,
28,
29]. The removal of poorly segmented trees was necessary to ensure the quality of the extracted trees, but drastically reduced the number of analyzed trees. This type of manual removal of trees results in the methods presented here being unable to be applied across most forest stands. Therefore, improvements in the quality and reliability of automatic tree segmentation algorithms should be seen as a prerequisite to the broader applications of individual-tree-based work. A collaborative approach between multi-disciplinary researchers is likely required. The authors support the creation of an open dataset of point clouds that could be used to further develop of these algorithms. Ideally, such a dataset would include various types of segmented and non-segmented point clouds collected from a range of forest types.
It is important to note that the volumes reported in this study do not necessarily represent the true volume of each live branch cluster or individual tree. This work instead relied on automated methods and manual measurements that assumed a tree was composed of a series of cylinders of various vertical lengths and diameters. This was done primarily to allow for manual measurements to be used for validation without having to destructively sample trees. While these methods are an improvement to existing field-based method, they likely overestimate the true volume of each live branch cluster and therefore the total volume. In reality, these volumes vary depending on how detailed the shapes are that are used to measure them, with finer detailed shapes providing more accurate volume estimates. Therefore, we suggest future research follow similar methods but use an approach that calculates the three-dimensional volume of each cluster using detailed alpha shapes. This type of work could then be used to improve conventional fire behaviour and fuel models by pairing these methods with destructive sampling-based techniques. Such methods could be used to develop novel, fine-scale allometric models that could predict the weight of the fuel within each cluster. This could be used along with the cluster volumes to extract inner-tree crown bulk density measurements. These fine-scale measurements could then be used to provide more accurate stand-level canopy bulk density measurements. Additionally, the fine-scale of the metrics extracted by these methods suggest that they could be used to build and further develop next-generation fire behaviour models, such as those described in [
41].