Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds
Abstract
:1. Introduction
2. Materials and Methods
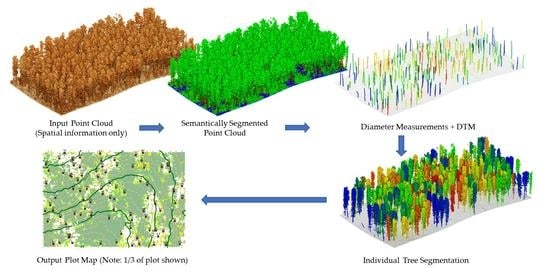
2.1. Forest Structural Complexity Tool Overview
2.1.1. Point Cloud Semantic Segmentation
2.1.2. Digital Terrain Model
2.1.3. Point Cloud Cleaning after Segmentation
2.1.4. Stem Point Cloud Skeletonization
2.1.5. Skeleton Clustering into Branch/Stem Segments
2.1.6. Cylinder Fitting
2.1.7. Sorting Cylinder Measurements into Individual Trees
| Algorithm 1. Cylinder Sorting Algorithm Part 1. |
For clarity, we will label a variable “TREE_ID” as uppercase and the tree_id belonging to a cylinder point as “assigned_tree_id”.
Loop until unsorted_points is empty.
|
| Algorithm 2. Cylinder Sorting Algorithm Part 2 |
|
2.1.8. Cylinder Measurement Interpolation
2.1.9. Cylinder Measurement Smoothing and Cleaning
2.1.10. Stem Volume Extraction
2.1.11. Individual Tree Segmentation of Vegetation and Stem points

2.1.12. Automated Height Measurement Extraction
2.1.13. Automated Diameter at Breast Height (DBH) Measurement Extraction
2.2. Reference Data Collection—Destructively Sampled Manual Field Measurements
2.3. Data Collection—Terrestrial Laser Scanning of Plots
2.4. Validation Process—Comparing Manual and Automated Point Cloud Measurements
2.4.1. Tree Matching
2.4.2. Taper Measurement Matching
2.4.3. Reference Volume
2.4.4. Plot Density
2.5. Qualitative Demonstration of FSCT on 5 Sensor/Structure Diverse Point Clouds
2.6. Computer Hardware Used for Run times
3. Results
3.1. Diameter at Breast Height (DBH)
3.2. Tree Height
3.3. All Stem Diameter Measurements
3.4. Tree and Measurement Detection Completeness

3.5. Stem Volume
3.6. Stem Density Estimates
3.7. Run Times
3.8. Video Demonstration of FSCT on Other Point Cloud Datasets
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Plot | # Ref. Trees | # Trees Detected | Tree Detection Completeness | # Ref. Diameter Measurements | # Matched Diameter Measurements | Diameter Measurement Completeness | Mean Diameter Error (m) | Root Mean Squared Error of Diameter (m) |
|---|---|---|---|---|---|---|---|---|
| 1 | 12 | 12 | 1.00 | 158 | 148 | 0.94 | 0.074 | 0.120 |
| 2 | 12 | 12 | 1.00 | 151 | 110 | 0.73 | 0.057 | 0.102 |
| 3 | 12 | 12 | 1.00 | 187 | 140 | 0.75 | 0.006 | 0.106 |
| 4 | 12 | 10 | 0.83 | 150 | 107 | 0.71 | 0.013 | 0.112 |
| 5 | 12 | 12 | 1.00 | 177 | 140 | 0.79 | 0.004 | 0.128 |
| 6 | 12 | 12 | 1.00 | 228 | 186 | 0.82 | 0.048 | 0.140 |
| 7 | 12 | 12 | 1.00 | 248 | 201 | 0.81 | 0.048 | 0.154 |
| 8 | 12 | 11 | 0.92 | 189 | 160 | 0.85 | 0.067 | 0.131 |
| 9 | 12 | 12 | 1.00 | 190 | 156 | 0.82 | 0.048 | 0.137 |
| 10 | 12 | 12 | 1.00 | 86 | 60 | 0.70 | 0.006 | 0.046 |
| 11 | 12 | 12 | 1.00 | 72 | 60 | 0.83 | 0.018 | 0.040 |
| 12 | 12 | 12 | 1.00 | 78 | 56 | 0.72 | 0.006 | 0.028 |
| 13 | 12 | 12 | 1.00 | 124 | 73 | 0.59 | 0.034 | 0.085 |
| 14 | 12 | 12 | 1.00 | 87 | 66 | 0.76 | 0.003 | 0.048 |
| 15 | 12 | 11 | 0.92 | 96 | 60 | 0.63 | −0.001 | 0.032 |
| 16 | 12 | 12 | 1.00 | 150 | 103 | 0.69 | 0.033 | 0.106 |
| 17 | 12 | 12 | 1.00 | 131 | 104 | 0.79 | 0.053 | 0.107 |
| 18 | 12 | 6 | 0.50 | 105 | 36 | 0.34 | 0.021 | 0.104 |
| 19 | 12 | 12 | 1.00 | 150 | 127 | 0.85 | 0.019 | 0.084 |
| 20 | 12 | 11 | 0.92 | 162 | 113 | 0.70 | 0.017 | 0.126 |
| 21 | 12 | 10 | 0.83 | 186 | 145 | 0.78 | 0.054 | 0.102 |
| 22 | 12 | 10 | 0.83 | 73 | 43 | 0.59 | 0.018 | 0.083 |
| 23 | 12 | 12 | 1.00 | 111 | 68 | 0.61 | 0.000 | 0.059 |
| 24 | 12 | 12 | 1.00 | 108 | 86 | 0.80 | 0.000 | 0.063 |
| 25 | 12 | 12 | 1.00 | 115 | 83 | 0.72 | 0.022 | 0.093 |
| 26 | 12 | 10 | 0.83 | 177 | 112 | 0.63 | 0.019 | 0.135 |
| 27 | 12 | 9 | 0.75 | 120 | 60 | 0.50 | −0.008 | 0.102 |
| Plot | # Ref. Trees | # Trees Detected | Tree Detection Completeness | # Ref. Diameter Measurements | # Matched Diameter Measurements | Diameter Measurement Completeness | Mean Diameter Error (m) | Root Mean Squared Error of Diameter (m) |
|---|---|---|---|---|---|---|---|---|
| 28 | 12 | 12 | 1.00 | 202 | 172 | 0.85 | 0.046 | 0.098 |
| 29 | 12 | 12 | 1.00 | 193 | 146 | 0.76 | 0.000 | 0.065 |
| 30 | 12 | 12 | 1.00 | 205 | 159 | 0.78 | 0.068 | 0.121 |
| 31 | 12 | 11 | 0.92 | 174 | 127 | 0.73 | 0.067 | 0.123 |
| 32 | 12 | 12 | 1.00 | 197 | 166 | 0.84 | 0.027 | 0.079 |
| 33 | 12 | 12 | 1.00 | 186 | 164 | 0.88 | 0.028 | 0.071 |
| 34 | 12 | 11 | 0.92 | 226 | 155 | 0.69 | 0.019 | 0.100 |
| 35 | 12 | 12 | 1.00 | 213 | 183 | 0.86 | 0.034 | 0.093 |
| 36 | 12 | 12 | 1.00 | 187 | 146 | 0.78 | 0.030 | 0.101 |
| 37 | 12 | 4 | 0.33 | 88 | 21 | 0.24 | −0.008 | 0.032 |
| 38 | 12 | 0 | 0.00 | 71 | 0 | 0.00 | 0.000 | 0.000 |
| 39 | 12 | 12 | 1.00 | 107 | 85 | 0.79 | 0.020 | 0.093 |
| 40 | 12 | 7 | 0.58 | 82 | 39 | 0.48 | 0.016 | 0.050 |
| 41 | 12 | 12 | 1.00 | 147 | 118 | 0.80 | 0.059 | 0.111 |
| 42 | 12 | 12 | 1.00 | 135 | 90 | 0.67 | 0.030 | 0.102 |
| 43 | 12 | 12 | 1.00 | 99 | 82 | 0.83 | 0.044 | 0.096 |
| 44 | 12 | 12 | 1.00 | 118 | 90 | 0.76 | 0.034 | 0.080 |
| 45 | 12 | 11 | 0.92 | 78 | 61 | 0.78 | 0.014 | 0.041 |
| 46 | 12 | 8 | 0.67 | 105 | 51 | 0.49 | 0.011 | 0.022 |
| 47 | 12 | 12 | 1.00 | 122 | 97 | 0.80 | 0.059 | 0.088 |
| 48 | 12 | 11 | 0.92 | 155 | 84 | 0.54 | 0.023 | 0.098 |
| 49 | 12 | 12 | 1.00 | 123 | 102 | 0.83 | 0.000 | 0.062 |
Appendix B. Qualitative Demonstration Video Notes
References
- Holopainen, M.; Kankare, V.; Vastaranta, M.; Liang, X.; Lin, Y.; Vaaja, M.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Kaartinen, H.; et al. Tree mapping using airborne, terrestrial and mobile laser scanning—A case study in a heterogeneous urban forest. Urban For. Urban Green. 2013, 12, 546–553. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A low-cost multi-sensoral mobile mapping system and its feasibility for tree measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- Kukko, A.; Kaijaluoto, R.; Kaartinen, H.; Lehtola, V.V.; Jaakkola, A.; Hyyppä, J. Graph SLAM correction for single scanner MLS forest data under boreal forest canopy. ISPRS J. Photogramm. Remote Sens. 2017, 132 (Suppl. C), 199–209. [Google Scholar] [CrossRef]
- Wang, J.; Lindenbergh, R. Validating a workflow for tree inventory updating with 3d point clouds obtained by mobile laser scanning. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, 42, 2. [Google Scholar] [CrossRef] [Green Version]
- Piermattei, L.; Karel, W.; Wang, D.; Wieser, M.; Mokroš, M.; Surový, P.; Koreň, M.; Tomaštík, J.; Pfeifer, N.; Hollaus, M. Terrestrial Structure from Motion Photogrammetry for Deriving Forest Inventory Data. Remote Sens. 2019, 11, 950. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Jaakkola, A.; Wang, Y.; Hyyppä, J.; Honkavaara, E.; Liu, J.; Kaartinen, H. The Use of a Hand-Held Camera for Individual Tree 3D Mapping in Forest Sample Plots. Remote Sens. 2014, 6, 6587. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Wang, Y.; Jaakkola, A.; Kukko, A.; Kaartinen, H.; Hyyppä, J.; Honkavaara, E.; Liu, J. Forest Data Collection Using Terrestrial Image-Based Point Clouds from a Handheld Camera Compared to Terrestrial and Personal Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5117–5132. [Google Scholar] [CrossRef]
- Mokroš, M.; Liang, X.; Surový, P.; Valent, P.; Čerňava, J.; Chudý, F.; Tunák, D.; Saloň, Š.; Merganič, J. Evaluation of Close-Range Photogrammetry Image Collection Methods for Estimating Tree Diameters. ISPRS Int. J. Geo-Inf. 2018, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Kuželka, K.; Surový, P. Mapping Forest Structure Using UAS inside Flight Capabilities. Sensors 2018, 18, 2245. [Google Scholar] [CrossRef] [Green Version]
- Krisanski, S.; Taskhiri, M.S.; Turner, P. Enhancing Methods for Under-Canopy Unmanned Aircraft System Based Photogrammetry in Complex Forests for Tree Diameter Measurement. Remote Sens. 2020, 12, 1652. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest Inventory with Terrestrial LiDAR: A Comparison of Static and Hand-Held Mobile Laser Scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Bienert, A.; Scheller, S.; Keane, E.; Mohan, F.; Nugent, C. Tree detection and diameter estimations by analysis of forest terrestrial laserscanner point clouds. In Proceedings of the ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007. [Google Scholar]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, M.; Demol, M.; Disney, M.; et al. Terrestrial laser scanning in forest ecology: Expanding the horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Ghimire, S.; Xystrakis, F.; Koutsias, N. Using Terrestrial Laser Scanning to Measure Forest Inventory Parameters in a Mediterranean Coniferous Stand of Western Greece. PFG J. Photogramm. Remote Sens. Geoinf. Sci. 2017, 85, 213–225. [Google Scholar] [CrossRef]
- Jose, G.d.T.; Lau, A.; Bartholomeus, H.; Herold, M.; Avitabile, V.; Raumonen, P.; Martius, C.; Goodman, R.C.; Disney, M.; Manuri, S.; et al. Estimation of above-ground biomass of large tropical trees with terrestrial LiDAR. Methods Ecol. Evol. 2018, 9, 223–234. [Google Scholar]
- Krishna Moorthy, S.M.; Raumonen, P.; Van den Bulcke, J.; Calders, K.; Verbeeck, H. Terrestrial laser scanning for non-destructive estimates of liana stem biomass. For. Ecol. Manag. 2020, 456, 117751. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Moskal, L.M.; Zheng, G. Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest. Remote Sens. 2012, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Danson, M. Terrestrial Laser Scanning for Plot-Scale Forest Measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Saarinen, N.; Luoma, V.; Tanhuanpää, T.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Detecting and characterizing downed dead wood using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2019, 151, 76–90. [Google Scholar] [CrossRef]
- Stephenson, P.J. Integrating Remote Sensing into Wildlife Monitoring for Conservation. Environ. Conserv. 2019, 46, 181–183. [Google Scholar] [CrossRef]
- McElhinny, C.; Gibbons, P.; Brack, C. An objective and quantitative methodology for constructing an index of stand structural complexity. For. Ecol. Manag. 2006, 235, 54–71. [Google Scholar] [CrossRef]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J. Forest and woodland stand structural complexity: Its definition and measurement. For. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Breshears, D.D.; Myers, O.B.; Meyer, C.W.; Barnes, F.J.; Zou, C.B.; Allen, C.D.; McDowell, N.G.; Pockman, W.T. Tree die-off in response to global change-type drought: Mortality insights from a decade of plant water potential measurements. Front. Ecol. Environ. 2009, 7, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Brown, S. Measuring carbon in forests: Current status and future challenges. Environ. Pollut. 2002, 116, 363–372. [Google Scholar] [CrossRef]
- Csillik, O.; Kumar, P.; Mascaro, J.; O’Shea, T.; Asner, G.P. Monitoring tropical forest carbon stocks and emissions using Planet satellite data. Sci. Rep. 2019, 9, 17831. [Google Scholar] [CrossRef] [Green Version]
- Novo, A.; Fariñas-Álvarez, N.; Martínez-Sánchez, J.; González-Jorge, H.; Fernández-Alonso, J.M.; Lorenzo, H. Mapping Forest Fire Risk—A Case Study in Galicia (Spain). Remote Sens. 2020, 12, 3705. [Google Scholar] [CrossRef]
- DeFries, R. Why forest monitoring matters for people and the planet. In Global Forest Monitoring from Earth Observation; CRC Press: Boca Raton, FL, USA, 2013; pp. 1–14. [Google Scholar]
- Dubayah, R.; Blair, K.B.; Goetz, S.; Fatoyinbo, L.; Hansen, M.; Healey, S.; Hofton, M.; Hurtt, G.; Kellner, J.; Luthcke, S.; et al. The Global Ecosystem Dynamics Investigation: High-resolution laser ranging of the Earth’s forests and topography. Sci. Remote Sens. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Johansen, K.; Phinn, S.; Taylor, M. Mapping woody vegetation clearing in Queensland, Australia from Landsat imagery using the Google Earth Engine. Remote Sens. Appl. Soc. Environ. 2015, 1, 36–49. [Google Scholar] [CrossRef]
- Piboule, A.; Krebs, M.; Esclatine, L.; Hervé, J.C. Computree: A collaborative platform for use of terrestrial lidar in dendrometry. In Proceedings of the International IUFRO Conference 2013: MeMoWood, Nancy, France, 1–4 October 2013. [Google Scholar]
- Burt, A.; Disney, M.; Calders, K. Extracting individual trees from lidar point clouds using treeseg. Methods Ecol. Evol. 2018, 10, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Takoudjou, S.M.; Casella, E. LeWoS: A universal leaf-wood classification method to facilitate the 3D modelling of large tropical trees using terrestrial LiDAR. Methods Ecol. Evol. 2020, 11, 376–389. [Google Scholar] [CrossRef]
- Wang, D. Unsupervised semantic and instance segmentation of forest point clouds. ISPRS J. Photogramm. Remote Sens. 2020, 165, 86–97. [Google Scholar] [CrossRef]
- Morel, J.; Bac, A.; Kanai, T. Segmentation of unbalanced and in-homogeneous point clouds and its application to 3D scanned trees. Vis. Comput. 2020, 36, 2419–2431. [Google Scholar] [CrossRef]
- Raumonen, P.; Casella, E.; Calders, K.; Murphy, S.; Akerblom, M.; Kaasalainen, M. Massive-scale tree modelling from TLS data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 2, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Marselis, S.; Yebra, M.; Jovanovic, T.; Ide Jan Martijn van Dijk, A. Deriving comprehensive forest structure information from mobile laser scanning observations using automated point cloud classification. Environ. Model. Softw. 2016, 82, 142–151. [Google Scholar] [CrossRef]
- Windrim, L.; Bryson, M. Detection, Segmentation, and Model Fitting of Individual Tree Stems from Airborne Laser Scanning of Forests Using Deep Learning. Remote Sens. 2020, 12, 1469. [Google Scholar] [CrossRef]
- Heinzel, J.; Huber, M.O. Detecting Tree Stems from Volumetric TLS Data in Forest Environments with Rich Understory. Remote Sens. 2017, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Koreň, M. DendroCloud: Point Cloud Processing Software for Forestry; Technical University in Zvolen: Zvolen, Slovakia, 2018. [Google Scholar]
- GreenValley International, LIDAR360 Comprehensive Point Cloud Post-Processing Suite. 2020. Available online: https://greenvalleyintl.com/software/lidar360/ (accessed on 16 June 2021).
- Trochta, J.; Krůček, M.; Vrška, T.; Král, K. 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE 2017, 12, e0176871. [Google Scholar] [CrossRef] [Green Version]
- Lalonde, J.-F.; Vandapel, N.; Hebert, M. Automatic Three-Dimensional Point Cloud Processing for Forest Inventory; The Robotics Institute, Carnegie Mellon University: Pittsburgh, PA, USA, 2006. [Google Scholar]
- Ayrey, E.; Fraver, S.; Kershaw Jr., J. A.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A Novel Algorithm for Individual Forest Tree Segmentation from LiDAR Point Clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Ayrey, E.; Hayes, D.J. The Use of Three-Dimensional Convolutional Neural Networks to Interpret LiDAR for Forest Inventory. Remote Sens. 2018, 10, 649. [Google Scholar] [CrossRef] [Green Version]
- Fan, G.; Nan, L.; Dong, Y.; Su, X.; Chen, F. AdQSM: A New Method for Estimating Above-Ground Biomass from TLS Point Clouds. Remote Sens. 2020, 12, 3089. [Google Scholar] [CrossRef]
- Delagrange, S.; Jauvin, C.; Rochon, P. PypeTree: A Tool for Reconstructing Tree Perennial Tissues from Point Clouds. Sensors 2014, 14, 4271–4289. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree—An Efficient Open Source Tool to Build Tree Models from TLS Clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Yan, D.; Wintz, J.; Mourrain, B.; Wang, W.; Boudon, F.; Godin, C. Efficient and robust reconstruction of botanical branching structure from laser scanned points. In Proceedings of the 2009 11th IEEE International Conference on Computer-Aided Design and Computer Graphics, Huangshan, China, 19–21 August 2009. [Google Scholar]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491. [Google Scholar] [CrossRef] [Green Version]
- Stovall, A.E.L.; Vorster, A.G.; Anderson, R.S.; Evangelista, P.H.; Shugart, H. Non-destructive aboveground biomass estimation of coniferous trees using terrestrial LiDAR. Remote Sens. Environ. 2017, 200, 31–42. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; Rex, F.E.; Almeida, D.R.A.d.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Zambrano, A.M.A.; Cunha Neto, E.M.d.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef] [Green Version]
- Aijazi, A.K.; Checchin, P.; Malaterre, L.; Trassoudaine, L. Automatic Detection and Parameter Estimation of Trees for Forest Inventory Applications Using 3D Terrestrial LiDAR. Remote Sens. 2017, 9, 946. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. arXiv 2017, arXiv:1706.02413. [Google Scholar]
- Krisanski, S.; Taskhiri, M.S.; Gonzalez Aracil, S.; Herries, D.; Turner, P. Sensor Agnostic Semantic Segmentation of Structurally Diverse and Complex Forest Point Clouds Using Deep Learning. Remote Sens. 2021, 13, 1413. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Astels, S. Hdbscan: Hierarchical density based clustering. J. Open Source Softw. 2017, 2, 205. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise; Institute for Computer Science, University of Munich: Munich, Germany, 1996. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Google. Map Showing Location of Study Site. Google Earth. 2019. Available online: https://earth.google.com (accessed on 16 June 2021).
- Liu, G.; Wang, J.; Dong, P.; Chen, Y.; Liu, Z. Estimating Individual Tree Height and Diameter at Breast Height (DBH) from Terrestrial Laser Scanning (TLS) Data at Plot Level. Forests 2018, 9, 398. [Google Scholar] [CrossRef] [Green Version]
- Ojoatre, S.; Zhang, C.; Hussin, Y.A.; Kloosterman, H.E.; Hasmadi Ismail, M. Assessing the Uncertainty of Tree Height and Aboveground Biomass From Terrestrial Laser Scanner and Hypsometer Using Airborne LiDAR Data in Tropical Rainforests. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 4149–4159. [Google Scholar] [CrossRef] [Green Version]
- Vasilescu, M.M. Standard error of tree height using Vertex III. Bull. Transilv. Univ. Brasov. For. Wood Ind. Agric. Food Eng. Ser. II 2013, 6, 76–80. [Google Scholar]

















| Western Australia | Sampled Tree Statistics (12 Trees per Plot) | ||||||
|---|---|---|---|---|---|---|---|
| Plot ID | Min DBH (m) | Mean DBH (m) | Max DBH (m) | Min Height (m) | Mean Height (m) | Max Height (m) | Stems/Ha |
| 1 | 0.063 | 0.207 | 0.276 | 10 | 19 | 23 | 725 |
| 2 | 0.08 | 0.179 | 0.271 | 14 | 18 | 21 | 575 |
| 3 | 0.075 | 0.200 | 0.283 | 12 | 23 | 26 | 725 |
| 4 | 0.056 | 0.181 | 0.313 | 9 | 18 | 25 | 700 |
| 5 | 0.126 | 0.204 | 0.305 | 15 | 21 | 25 | 650 |
| 6 | 0.117 | 0.248 | 0.338 | 21 | 28 | 33 | 500 |
| 7 | 0.134 | 0.269 | 0.398 | 24 | 30 | 34 | 575 |
| 8 | 0.119 | 0.185 | 0.282 | 19 | 24 | 29 | 1150 |
| 9 | 0.107 | 0.199 | 0.286 | 16 | 24 | 29 | 900 |
| 10 | 0.071 | 0.120 | 0.184 | 8 | 11 | 14 | 775 |
| 11 | 0.035 | 0.083 | 0.148 | 5 | 8 | 12 | 775 |
| 12 | 0.071 | 0.094 | 0.114 | 7 | 9 | 10 | 1000 |
| 13 | 0.065 | 0.152 | 0.234 | 9 | 17 | 20 | 1100 |
| 14 | 0.036 | 0.124 | 0.208 | 6 | 12 | 16 | 1025 |
| 15 | 0.073 | 0.125 | 0.173 | 8 | 11 | 14 | 775 |
| 16 | 0.084 | 0.183 | 0.292 | 12 | 19 | 24 | 925 |
| 17 | 0.063 | 0.128 | 0.203 | 11 | 16 | 20 | 1000 |
| 18 | 0.052 | 0.130 | 0.242 | 7 | 12 | 19 | 600 |
| 19 | 0.062 | 0.170 | 0.259 | 8 | 18 | 25 | 650 |
| 20 | 0.065 | 0.184 | 0.341 | 10 | 19 | 31 | 500 |
| 21 | 0.130 | 0.193 | 0.279 | 17 | 22 | 26 | 625 |
| 22 | 0.024 | 0.091 | 0.173 | 3 | 10 | 14 | 1425 |
| 23 | 0.061 | 0.132 | 0.209 | 8 | 14 | 18 | 975 |
| 24 | 0.062 | 0.121 | 0.179 | 8 | 14 | 18 | 1575 |
| 25 | 0.057 | 0.141 | 0.187 | 9 | 16 | 20 | 775 |
| 26 | 0.076 | 0.191 | 0.293 | 12 | 22 | 32 | 750 |
| 27 | 0.043 | 0.125 | 0.243 | 6 | 15 | 23 | 1075 |
| Green Triangle | Sampled Tree Statistics (12 Trees per Plot) | ||||||
|---|---|---|---|---|---|---|---|
| Plot ID | Min DBH (m) | Mean DBH (m) | Max DBH (m) | Min Height (m) | Mean Height (m) | Max Height (m) | Stems/Ha |
| 28 | 0.093 | 0.228 | 0.366 | 15 | 26 | 31 | 600 |
| 29 | 0.136 | 0.220 | 0.304 | 21 | 25 | 28 | 875 |
| 30 | 0.150 | 0.206 | 0.270 | 20 | 24 | 28 | 775 |
| 31 | 0.088 | 0.220 | 0.331 | 14 | 22 | 26 | 625 |
| 32 | 0.162 | 0.232 | 0.310 | 20 | 25 | 29 | 725 |
| 33 | 0.134 | 0.215 | 0.297 | 19 | 24 | 27 | 750 |
| 34 | 0.115 | 0.233 | 0.319 | 19 | 30 | 36 | 500 |
| 35 | 0.122 | 0.237 | 0.342 | 20 | 28 | 32 | 650 |
| 36 | 0.130 | 0.196 | 0.288 | 20 | 24 | 29 | 750 |
| 37 | 0.051 | 0.121 | 0.171 | 6 | 13 | 28 | 975 |
| 38 | 0.059 | 0.090 | 0.125 | 7 | 8 | 9 | 800 |
| 39 | 0.057 | 0.135 | 0.204 | 8 | 14 | 18 | 1000 |
| 40 | 0.070 | 0.123 | 0.185 | 6 | 10 | 12 | 725 |
| 41 | 0.084 | 0.183 | 0.266 | 12 | 19 | 23 | 625 |
| 42 | 0.107 | 0.168 | 0.263 | 14 | 17 | 20 | 675 |
| 43 | 0.05 | 0.113 | 0.201 | 8 | 13 | 19 | 1050 |
| 44 | 0.081 | 0.133 | 0.177 | 10 | 14 | 16 | 800 |
| 45 | 0.054 | 0.096 | 0.132 | 8 | 10 | 12 | 1000 |
| 46 | 0.063 | 0.129 | 0.192 | 9 | 12 | 15 | 775 |
| 47 | 0.074 | 0.169 | 0.260 | 9 | 16 | 20 | 750 |
| 48 | 0.072 | 0.175 | 0.273 | 12 | 20 | 25 | 800 |
| 49 | 0.064 | 0.147 | 0.215 | 6 | 16 | 22 | 850 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krisanski, S.; Taskhiri, M.S.; Gonzalez Aracil, S.; Herries, D.; Muneri, A.; Gurung, M.B.; Montgomery, J.; Turner, P. Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds. Remote Sens. 2021, 13, 4677. https://doi.org/10.3390/rs13224677
Krisanski S, Taskhiri MS, Gonzalez Aracil S, Herries D, Muneri A, Gurung MB, Montgomery J, Turner P. Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds. Remote Sensing. 2021; 13(22):4677. https://doi.org/10.3390/rs13224677
Chicago/Turabian StyleKrisanski, Sean, Mohammad Sadegh Taskhiri, Susana Gonzalez Aracil, David Herries, Allie Muneri, Mohan Babu Gurung, James Montgomery, and Paul Turner. 2021. "Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds" Remote Sensing 13, no. 22: 4677. https://doi.org/10.3390/rs13224677
APA StyleKrisanski, S., Taskhiri, M. S., Gonzalez Aracil, S., Herries, D., Muneri, A., Gurung, M. B., Montgomery, J., & Turner, P. (2021). Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds. Remote Sensing, 13(22), 4677. https://doi.org/10.3390/rs13224677