Forest Structural Estimates Derived Using a Practical, Open-Source Lidar-Processing Workflow
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Forestry Plot Data
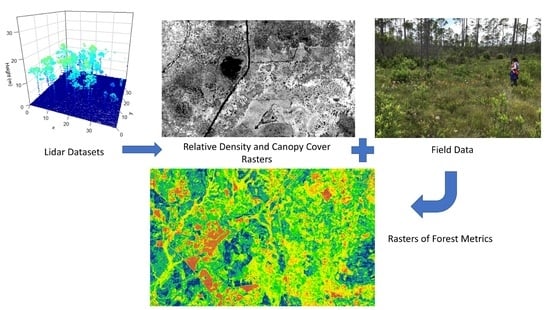
2.3. Lidar Data
2.4. Relative Density Canopy Cover Function
2.5. Cloud Processing
2.6. Forest Metric Models and Raster Outputs
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B









References
- Beland, M.; Geoffrey, P.; Sparrow, B.; Harding, D.; Chasmer, L.; Phinn, S.; Antonarakis, A.; Strahler, A. On promoting the use of lidar systems in forest ecosystem research. For. Ecol. Manag. 2019, 450, 117484. [Google Scholar] [CrossRef]
- USGS 3D Elevation Program. Available online: https://www.usgs.gov/core-science-systems/ngp/3dep (accessed on 1 August 2019).
- Meijer, C.; Grootes, M.; Koma, Z.; Dzigan, Y.; Gonçalves, R.; Andela, B.; van den Oord, G.; Ranguelova, E.; Renaud, N.; Kissling, W. Laserchicken-A tool for distributed feature calculation from massive Lidar point cloud datasets. SoftwareX 2020, 12, 100626. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne Lidar data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Drake, J.B.; Knox, R.G.; Dubayah, R.O.; Clark, D.B.; Condit, R.; Blair, J.B.; Hofton, M. Above-ground biomass estimation in closed canopy Neotropical forests using lidar remote sensing: Factors affecting the generality of relationships. Glob. Ecol. Biogeogr. 2003, 12, 147–159. [Google Scholar] [CrossRef]
- Pearse, G.D.; Dash, J.P.; Persson, H.J.; Watt, M.S. Comparison of high-density Lidar and satellite photogrammetry for forest inventory. ISPRS J. Photogramm. Remote Sens. 2018, 142, 257–267. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree Species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform Lidar data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Jarron, L.R.; Coops, N.C.; MacKenzie, W.H.; Tompalski, P.; Dykstra, P. Detection of sub-canopy forest structure using airborne Lidar. Remote Sens. Environ. 2020, 244, 111770. [Google Scholar] [CrossRef]
- Ahl, R.; Hogland, J.; Brown, S. A Comparison of Standard Modeling Techniques Using Digital Aerial Imagery with National Elevation Datasets and Airborne Lidar to Predict Size and Density Forest Metrics in the Sapphire Mountains MT, USA. Int. J. Geo.-Inf. 2019, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Nordman, C.; White, R.; Wilson, R.; Ware, C.; Rideout, C.; Pyne, M.; Hunter, C. Rapid Assessment Metrics to Enhance Wildlife Habitat and Biodiversity within Southern Open Pine Ecosystems; U.S. Fish and Wildlife Service and NatureServe, for the Gulf Coastal Plains and Ozarks Landscape Conservation Cooperative: Tallahassee, FL, USA, 2016. [Google Scholar]
- Trager, M.D.; Drake, J.B.; Jenkins, A.M.; Petrick, C.J. Mapping and Modeling Ecological Conditions of Longleaf Pine Habitats in the Apalachicola National Forest. For. Ecol. 2018, 116, 304–311. [Google Scholar] [CrossRef]
- Garabedian, J.; Moorman, C.; Peterson, M.N.; Kilgo, J. Use of Lidar to define habitat thresholds for forest bird conservation. For. Ecol. Manag. 2017, 399, 24–36. [Google Scholar] [CrossRef]
- Cohen, M.; McLaughlin, D.; Kaplan, D.; Acharya, S. Managing Forest for Increase Regional Water Availability; Florida Department of Agriculture and Consumer Services: Tallahassee, FL, USA, 2017. Available online: https://www.fdacs.gov/content/download/76293/file/20834_Del_7.pdf (accessed on 9 August 2021).
- Darracq, A.K.; Boone, W.W.; McCleery, R.A. Burn regime matters: A review of the effects of prescribed fire on vertebrates in the longleaf pine ecosystem. For. Ecol. Manag. 2016, 378, 214–221. [Google Scholar] [CrossRef]
- Young, J.A.; Mahan, C.G.; Forder, M. Integration of Vegetation Community Spatial Data into a Prescribed Fire Planning Process at Shenandoah National Park Virginia (USA). Nat. Areas J. 2017, 37, 394–405. [Google Scholar] [CrossRef]
- Zald, H.S.; Wulder, M.A.; White, J.C.; Hilker, T.; Hermosilla, T.; Hobart, G.W.; Coops, N.C. Integrating Landsat pixel composites and changemetrics with lidar plots to predictively map forest structure and aboveground biomass in Saskatchewan, Canada. Remote Sens. Environ. 2015, 176, 188–201. [Google Scholar] [CrossRef] [Green Version]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Isenburg, M. LASzip: Lossless compression of Lidar data. Photogramm. Eng. Remote Sens. 2013, 79, 209–217. [Google Scholar] [CrossRef]
- Lindsay, J.; Whitebox Geospatial Inc. Whitebox Geo. 2021. Available online: https://www.whiteboxgeo.com/ (accessed on 17 September 2021).
- McGaughey, R.J. FUSION/LDV LIDAR Analysis and Visualization Software. Pacific Northwest Research Station USDA Fortest Service. Available online: http://forsys.cfr.washington.edu/FUSION/fusion_overview.html (accessed on 1 September 2019).
- Silva, C.A.; Crookston, N.L.; Hudak, A.T.; Vierling, L.A.; Klauberg, C.; Cardil, A.; Hamamura, C. rLidar: An R Package for Reading, Processing and Visualizing Lidar (Light Detection and Ranging) Data. R Package Version 0.1.5. 2021. Available online: https://cran.r-project.org/web/packages/rLidar/index.html (accessed on 18 October 2021).
- Roussel, J.; Auty, D.; Coops, N.; Tompalski, P.; Goodbody, T.; Meador, A.; Bourdon, J.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- Stein, B.A.; Kutner, L.S.; Adams, J.S. Precious Heritage: The Status of Biodiversity in the United States; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- U.S. Department of Agriculture. USDA RESTORE Council to Invest 31 million for Priority Restoration Work. U.S. Dep. Agric. Press Releases. Available online: https://www.usda.gov/media/press-releases/2021/04/29/usda-restore-council-invest-31-million-priority-restoration-work (accessed on 10 May 2021).
- Noss, R.F. Longleaf pine and wiregrass: Keystone components of an endangered Ecosystem. Nat. Areas J. 1989, 9, 211–213. [Google Scholar]
- Florida Sea Grant. Apalachicola Bay Oyster Situation Report (TP-200). 2013. Available online: https://www.flseagrant.org/wp-content/uploads/tp200_apalachicola_oyster_situation_report.pdf (accessed on 10 March 2020).
- Hogland, J.; Affleck, D.L.; Anderson, N.; Seielstad, C.; Dobrowski, S.; Graham, J.; Smith, R. Estimating Forest Characteristics for Longleaf Pine. Forests 2020, 11, 426. [Google Scholar] [CrossRef]
- Florida Natural Areas Inventory. Natural Communities Guide; FNAI: Tallahassee, FL, USA. Available online: https://www.fnai.org/naturalcommguide.cfm (accessed on 24 June 2021).
- OCM Partners. 2018 TLCGIS Lidar: Leon County, FL. NOAA Fisheries. Available online: https://www.fisheries.noaa.gov/inport/item/60045 (accessed on 25 June 2021).
- OCM Partners. 2018 TLCGIS Lidar: Florida Panhandle. NOAA Fisheries. Available online: https://www.fisheries.noaa.gov/inport/item/58298 (accessed on 25 June 2021).
- OCM Partners. 2017 NWFWMD Lidar: Lower Choctawhatchee. NOAA Fisheries. Available online: https://www.fisheries.noaa.gov/inport/item/55725 (accessed on 25 June 2021).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 25 June 2021).
- Roussel, J.; Auty, D. Airborne Lidar Data Manipulation and Visualization for Forestry Applications. R Package Version 3.1.4. 2021. Available online: https://cran.r-project.org/package=lidR (accessed on 25 June 2021).
- Brian, G.P.; Peter, C. Performance Analytics: Econometric Tools for Performance and Risk Analysis. R Package Version 2.0.4. 2020. Available online: https://CRAN.R-project.org/package=PerformanceAnalytics (accessed on 25 June 2021).
- Wei, T.; Simko, V. Corrplot: Visualization of a Correlation Matrix. R Package Version 0.88. 2021. Available online: https://github.com/taiyun/corrplot (accessed on 25 June 2021).
- Harrell, F.E., Jr.; Dupont, C. Hmisc: Harrell Miscellaneous. R Package Version 4.5-0. 2021. Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 25 June 2021).
- American Society for Photogrammetry & Remote Sensing. LAS Specification 1.4-R15; ASPRS The Imaging & Geospatial Information Society: Bethesda, MD, USA, 2019. [Google Scholar]
- Hogland, J.; Anderson, N. Function Modeling Improves the Efficiency of Spatial Modeling Using Big Data from Remote Sensing. Big Data Cogn. Comput. 2017, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Hogland, J.; Anderson, N.; Affleck, D.L.; St. Peter, J. Using Forest Inventory Data with Landsat 8 Imagery to Map Longleaf Pine Forest Characteristics in Georgia, USA. Remote Sens. 2019, 11, 1803. [Google Scholar] [CrossRef] [Green Version]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- St. Peter, J.; Anderson, C.; Drake, J.; Medley, P. Spatially Quantifying Forest Loss at Landscape-scale Following a Major Storm Event. Remote Sens. 2020, 12, 1138. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Huang, Q. Spatial Cloud Computing: A Practical Approach; CRC Press: Bacon Raton, FL, USA, 2013. [Google Scholar]
- Da Silva, V.S.; Silva, C.A.; Silva, E.A.; Klauberg, C.; Mohan, M.; Dias, I.M.; Rex, F.E.; Loureiro, G.H. Effects of Modeling Methods and Sample Size for Lidar-Derived Basal Area Estimation in Eucalyptus Forest. In Proceedings of the Anais do XIX Simpósio Brasileiro de Sensoriamento Remoto, São José dos Campos, Brazil, 2019; Available online: https://proceedings.science/sbsr-2019/papers/effects-of-modeling-methods-and-sample-size-for-lidar-derived-basal-area-estimation-in-eucalyptus-forest?lang=en (accessed on 25 June 2021).
- Woods, M.; Lim, K.; Treitz, P. Predicting Forest stand variables from Lidar data in the Great Lakes-St. Lawrence forest of Ontario. For. Chron. 2008, 84, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Van Ewijk, K.Y.; Treitz, P.M.; Scott, N.A. Characterizing Forest Succession in Central Ontario using Lidar-derived Indices. Photogramm. Eng. Remote Sens. 2011, 77, 261–269. [Google Scholar] [CrossRef]
- Van Lier, O.R.; Luther, J.E.; White, J.C.; Fournier, R.A.; Côté, J.-F. Effect of scan angle on ALS metrics and area-based predictions of forest attributes for balsam fir dominated stands. Forestry 2021, 1–24. [Google Scholar] [CrossRef]
- Dalla Corte, A.; Rex, F.; Almeida, D.; Sanquetta, C.; Silva, C.; Moura, M.; Wilkinson, B.; Almeyda Zombrano, A.M.; da Cunha Neto, E.M.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Leite, R.V.; Broadbent, E.N.; Jaafar, W.S.; Srinivasan, S.; Bajaj, S.; Dalla Corte, A.P.; do Amaral, C.H.; Gopan, G.; Saad, S.N.M.; et al. Individual tree detection using UAV-lidar and UAV-SfM data: A tutorial for beginners. Open Geosci. 2021, 13, 1028–1039. [Google Scholar] [CrossRef]





| BA (m2/ha) | TPH | QMD (cm) | Elevation (m) | |
|---|---|---|---|---|
| Min | 0 | 0 | 0 | 0.34 |
| Mean | 20.07 | 1032.95 | 16.72 | 15.11 |
| Median | 17.66 | 756.49 | 16.78 | 9.35 |
| Max | 76.87 | 5943.74 | 63.97 | 75.35 |
| SD | 16.11 | 179.55 | 8.61 | 15.17 |
| Band | Variable | Description |
|---|---|---|
| 1 | Num_Returns | Total number of returns in cell |
| 2 | Num_GrndRet | Number of ground returns in cell |
| 3 | Num_1stRet | Number of first returns in cell |
| 4 | Grnd_Elev | Ground elevations |
| 5 | Mn_RH | Mean of all relative heights |
| 6 | SD_RH | Std. dev of all relative heights |
| 7 | RHt_95th | Relative height 95% |
| 8 | RHt_90th | Relative height 90% |
| 9 | RHt_75th | Relative height 75% |
| 10 | RHt_50th | Relative height 50% |
| 11 | RHt_25th | Relative height 25% |
| 12 | RHt_10th | Relative height 10% |
| 13 | RHt_05th | Relative height 5% |
| 14 | RD_2to10ft | Relative density 0.6096-m to 3.048-m (shrubs) |
| 15 | RD_10to20ft | Relative density 3.048-m to 6.096-m (low midstory) |
| 16 | RD_20to49ft | Relative density 6.096-m to 14.935-m (high midstory) |
| 17 | RD_gt2ft | Relative density all returns ≥ 0.6096 m |
| 18 | RD_gt10ft | Relative density all returns ≥ 3.048 m |
| 19 | RD_gt20ft | Relative density all returns ≥ 6.096 m |
| 20 | RD_gt49ft | Relative density all returns ≥ 14.935 m |
| 21 | CC_gt2ft | Canopy cover ≥ 0.6096-m (based only on first returns) |
| 22 | CC_gt10ft | Canopy cover ≥ 3.048-m (based only on first returns) |
| 23 | CC_gt20ft | Canopy cover ≥ 6.096-m (based only on first returns) |
| 24 | CC_gt49ft | Canopy cover ≥ 14.935-m (based only on first returns) |
| 25 | MnRHgt2ft | Mean of all relative heights ≥ 0.6096 m |
| 26 | MnRHgt10ft | Mean of all relative heights ≥ 3.048 m |
| 27 | MnRHgt20ft | Mean of all relative heights ≥ 6.096 m |
| 28 | MnRHgt49ft | Mean of all relative heights ≥ 14.935 m |
| Response | Variables | Adj. R2 | v | RMSE | MAE | AIC |
|---|---|---|---|---|---|---|
| Sentinel-2 | 0.428 | 5 | 12.034 | 9.553 | 2660.07 | |
| BAH | RDCC | 0.774 | 4 | 7.590 | 5.118 | 2431.31 |
| Combo | 0.795 | 4 | 7.224 | 5.090 | 2406.99 | |
| Sentinel-2 | 0.203 | 7 | 36.201 | 23.895 | 1821.59 | |
| TPH | RDCC | 0.779 | 7 | 10.033 | 5.570 | 1505.89 |
| Combo | 0.773 | 6 | 10.353 | 5.618 | 1511.61 | |
| Sentinel-2 | 0.193 | 6 | 7.623 | 5.804 | 1254.85 | |
| QMD | RDCC | 0.593 | 5 | 5.428 | 3.747 | 1085.76 |
| Combo | 0.596 | 4 | 5.415 | 3.780 | 1082.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
St. Peter, J.; Drake, J.; Medley, P.; Ibeanusi, V. Forest Structural Estimates Derived Using a Practical, Open-Source Lidar-Processing Workflow. Remote Sens. 2021, 13, 4763. https://doi.org/10.3390/rs13234763
St. Peter J, Drake J, Medley P, Ibeanusi V. Forest Structural Estimates Derived Using a Practical, Open-Source Lidar-Processing Workflow. Remote Sensing. 2021; 13(23):4763. https://doi.org/10.3390/rs13234763
Chicago/Turabian StyleSt. Peter, Joseph, Jason Drake, Paul Medley, and Victor Ibeanusi. 2021. "Forest Structural Estimates Derived Using a Practical, Open-Source Lidar-Processing Workflow" Remote Sensing 13, no. 23: 4763. https://doi.org/10.3390/rs13234763
APA StyleSt. Peter, J., Drake, J., Medley, P., & Ibeanusi, V. (2021). Forest Structural Estimates Derived Using a Practical, Open-Source Lidar-Processing Workflow. Remote Sensing, 13(23), 4763. https://doi.org/10.3390/rs13234763