1. Introduction
Wheat is one of the three major food crops in China. Jiangsu Province, located in the lower reaches of the Yangtze River, is one of the major wheat production areas in China. In recent years, the delay in rice maturity in this region has led to a significant delay in subsequent winter wheat sowing. For example, the percentages of late-sown (seven or more days later than local normal seed sowing date) wheat area in Jiangsu Province were 48.6%, 51.2%, and 59.6% in 2015, 2016, and 2017, respectively [
1]. The delay in sowing has become a major obstacle to high wheat yield; therefore, it is necessary to screen wheat varieties suitable for late sowing. For example, optimal wheat varieties should be able to maintain a certain amount of growth even at the overwintering stage [
2]. In addition, wheat in the lower reach of the Yangtze River often suffers from low-temperature frost damage at the overwintering stage, which severely affects wheat growth and development [
3]; hence, good wheat varieties should have strong resistance to low temperatures. Therefore, accurately monitoring wheat growth status at the overwintering stage is critical for late-sown winter wheat variety screening.
As an important pigment for photosynthesis, chlorophyll has a critical impact on a plant’s ability to exchange material and energy with the external environment. Chlorophyll content can indicate crop growth status, primary productivity, and nitrogen use efficiency [
4]. Exposure to various kinds of stresses may reduce crop chlorophyll content [
5]; so, chlorophyll content can provide information on a wheat variety’s tolerance capability to endure stresses due to late sowing, low temperature, insufficient nitrogen application and so on.
The traditional methods for measuring chlorophyll content are usually time-consuming, laborious, and destructive to crop leaves [
4]. Although the Soil and Plant Analysis Development (SPAD) method is non-destructive, it can work only at limited measuring points, and cannot provide the spatially continuous distribution of SPAD [
6].
Late-sown wheat variety screening requires a large number of experimental plots. Better monitoring wheat growth status may benefit from rapid, accurate, and non-destructive estimation of wheat chlorophyll content in each plot. Remote sensing provides a great potential for chlorophyll estimation over large regions [
7].
Spectral vegetation indices (VIs) have been widely used to estimate vegetation chlorophyll content from spectral data [
8]. Good vegetation indices are able to maximize sensitivity to the vegetation characteristics, while reducing the spectral effects due to atmosphere, soil background, topography, and sensor view angle [
9,
10].
The VI of modified chlorophyll absorption in reflectance index (MCARI) that was proposed by Daughtry et al. [
11] is based on the reflectance of green, red, and red edge bands, and this VI is sensitive to leaf chlorophyll variations. Using the reflectance of green, red edge, and near-infrared (NIR) bands, Cao et al. [
12] modified MCARI to propose a new VI of modified chlorophyll absorption reflectance index 1 (MCARI1). They indicated that the MCARI1 displayed quite significant correlations with rice’s above-ground biomass and plant nitrogen uptake at each growth stage of rice.
The Medium Resolution Imaging Spectrometer (MERIS) terrestrial chlorophyll index (MTCI) that was designed by Dash and Curran [
13] is suitable for the estimation of chlorophyll content from the MERIS data. MTCI has become a frequently used VI to monitor spatial variability of crop chlorophyll [
14].
A 2-band VI of green chlorophyll vegetation indices (GCVI) that was proposed by Gitelson et al. [
15], using green and NIR bands, had good correlations with chlorophyll content in maize and soybean canopy. The developed models could provide accurate estimation of canopy chlorophyll contents, although calibration coefficients were different for maize and soybean.
The chlorophyll vegetation index (CVI) that was developed by Vincini et al. [
16] uses the reflectance of green, red, and NIR broad bands. Vincini et al. [
16] indicated that CVI was specifically sensitive to leaf chlorophyll content at the canopy scale of sugar beet.
Since hyperspectral data are composed of a large number of continuous and narrow bands, a number of spectral indices have been proposed to estimate the chlorophyll content of vegetation [
17]. Main et al. [
18] developed two red edge derivative based indices (i.e., red edge position via linear extrapolation index and the modified red edge inflection point index), and found that the two indices were consistent and robust in chlorophyll content estimation in three crop species and a variety of savanna tree species. Jin et al. [
19] developed a spectral index of double-peak canopy nitrogen index I (DCNI I), and they indicated that DCNI I produced accurate estimation of chlorophyll content in cotton. However, hyperspectral images are difficult to obtain. These hyperspectral VIs cannot be calculated from broadband multispectral data that are more readily available.
The spectral indices that were proposed for SPAD estimation use different spectral bands and have very different equations. Their performances vary among different studies, and none of them performed the best in all studies. More importantly, while most previous studies in remote sensing of SPAD only involved single variety or a very small number of varieties of a crop, very little research focused on the relationships between SPAD and spectral indices that were impacted by a large number of varieties. Accurate estimation of SPAD of various varieties is critical for late-sown wheat variety screening.
Traditional algorithms such as simple or multiple linear regressions have often been used in remote sensing for crop biophysical and biochemical variable retrieval. In recent years, machine learning algorithms (MLAs) have been employed increasingly. Different from traditional algorithms, MLAs are data-driven, and they are able to autonomously cope with linear correlations as well as solve strong nonlinear problems possessed by agricultural and remote sensing variables [
20].
Among a number of powerful MLAs, support vector regression (SVR), random forest regression (RFR), and Artificial Neural Network (ANN) are most frequently used for agricultural remote sensing [
21]. Yang et al. [
22] estimated green leaf chlorophyll density of rice from hyperspectral reflectance measured over two experimental rice fields containing two cultivars treated with three levels of nitrogen application. They found that SVR, the regression version of support vector machine (SVM), largely improved the estimation accuracy in comparison with the stepwise multiple regression (SMR). They indicated that SVR deals better than traditional regression algorithms with non-linear processes that exist in the relation between green leaf chlorophyll density and spectral data.
Similarly, an SVR-based model developed using the canopy spectral reflectance of maize, measured by a handheld spectrometer, was demonstrated to be able to estimate the chlorophyll content of maize canopy in the field non-destructively and rapidly [
23]. Cavallo et al. [
24] used RFR to predict total chlorophyll content of fresh-cut rocket leaves from spectral data measured using a spectrophotometer. The developed RFR model could provide accurate estimation of total chlorophyll content.
Although ANN is also widely used to estimate agricultural variables, it is not as practical as SVR and RFR because it requires complex and time consuming procedures such as selecting the number and size of hidden layers, setting the learning rate, obtaining a large training dataset, and dealing with the problem of overfitting [
20].
Prior to modeling, it is important to determine which variables should be included in the model, using a suitable variable selection method. A good variable selection method can select the smallest number and most efficient subset of variables from the original set, which can improve the estimation power of the model and speed up the model execution time [
25].
Random forest (RF) is not only a regression algorithm but also a frequently used variable selection method. This is because the importance of each variable can be calculated using RF and ranked during RF model development [
26]. Recursive feature elimination (RFE) is another widely used method for variable selection. The method uses a base model to perform multiple rounds of training, during which the weakest features are eliminated until a specified number of features is reached [
27,
28]. The base model could be SVM [
27], RF [
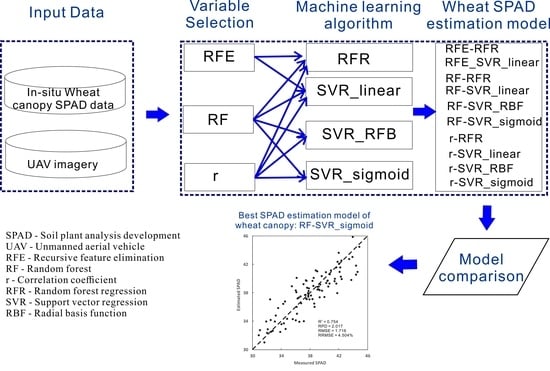
29], or other regression models. Nevertheless, there have been few previous studies that used variable selection in remote sensing of vegetation chlorophyll contents, although variable selection is critical. To fill this gap, the current study was designed to develop a good combination of variable selection methods with machine learning algorithms that can accurately estimate SPAD of wheat canopy at the overwintering stage, by comparative analysis. This study was based on the unmanned aerial vehicle (UAV) technology because the wheat variety screening experiments required high spatial resolution imagery.
4. Discussion
4.1. Optimal SPAD Estimation Model for Late-Sown Winter Wheat Variety Screening
This study involved up to 24 wheat varieties, four rates of nitrogen fertilization, and 96 plots in a variety screening experiment, which resulted in a very complex relationship between wheat canopy SPAD values and spectral variables. Therefore, this study investigated the feasibility of using machine learning-based models rather than conventional ones to estimate SPAD of wheat canopy from UAV data at the overwintering stage. Seven SVR models and three RFR models were developed, cross-validated, and compared. Of these 10 models, the RF-SVR_sigmoid model, which was constructed by combining the RF variable selection method and the sigmoid kernel-based SVR algorithm, was identified as the best model. It achieved the highest accuracy in estimating SPAD values by the LOOCV (R
2 = 0.754, RPD = 2.017, RMSE = 1.716, and RRMSE = 4.504%). According to Viscarra Rossel et al. [
67], the model of RF-SVR_sigmoid (RPD = 2.017) achieved very good SPAD estimation of wheat canopy. In contrast, r-SVR_sigmoid (RPD = 1.973) and RFE-SVR_linear (RPD = 1.926) only produced good estimation, but their RPD values were very close to 2.
Despite the fact that the study involved up to 24 different wheat varieties, the newly developed RF-SVR_sigmoid model was able to achieve high accuracy in SPAD estimation. Moreover, the model worked well for both plots under different rates of nitrogen fertilization and plots with springness, weak springness, and semi-winterness varieties. It is particularly important for wheat variety screening that the developed model can be used for all the wheat varieties involved, considering that the best varieties are usually screened from a large number of varieties. In contrast, most previous studies of SPAD remote sensing involved only a single variety or a very small number of varieties, which is not suitable for variety screening [
68,
69,
70].
4.2. Model Performance Comparison
A comparison of the seven SVR models (i.e., RFE-SVR_linear, RF-SVR_linear, RF-SVR_RBF, RF-SVR_sigmoid, r-SVR_linear, r-SVR_RBF, and r-SVR_sigmoid) with the three RFR models (i.e., RFE-RFR, RF-RFR, and r-RFR) finds that even the worst SVR model (i.e., r-SVR_linear) could produce better SPAD estimates (R2 = 0.710, RPD = 1.857, RMSE = 1.864, and RRMSE = 4.893%) than the best RFR model (i.e., RFE-RFR) (R2 = 0.640, RPD = 1.666, RMSE = 2.077, and RRMSE = 5.452%). In addition, the improvement in estimation accuracy was even greater when the best SVR model (i.e., the RF-SVR_sigmoid model) (R2 = 0.754, RPD = 2.017, RMSE = 1.716, and RRMSE = 4.504%) was compared with the best RFR model of RFE-RFR.
Given the encouraging performances of the SVR, however, it is too early to say that the SVR model always outperforms the RFR model in estimating vegetation chlorophyll content. For the estimation of other agricultural variables, some studies had reported that the RFR model outperformed the SVR model. For example, Osco et al. [
71] noted that, using UAV multispectral images, the RFR model was able to predict leaf nitrogen content (LNC) of maize more accurately than the SVR model. Similarly, Zha et al. [
65] reported that the RFR algorithm performed better than the SVR and ANN models in estimating the nitrogen nutrient index (NNI) of rice from drone data.
Yang et al. [
22] indicated that appropriate kernels could better avoid overfitting. The RBF kernel was considered to be the most frequently used kernel function [
72]. Some previous studies (e.g., Ahmad et al. [
73]; Chen and Hay [
74]) reported that the RBF kernel performed better relative to linear and sigmoid kernels. However, this study found that among the seven SVR models, the two sigmoid kernel-based SVR models performed the best, followed by the RFE-SVR_linear model, while the two RBF kernel-based SVR models produced lower accuracy. Hence, the sigmoid kernel seemed more appropriate for SVR in terms of SPAD estimation.
In addition, although the RFE-SVR_linear model was not as good as the two SVR models with sigmoid kernel, its SPAD estimation was also good. Moreover, the linear kernel–based SVR model runs much faster than the RBF kernel-based SVR model [
75].
4.3. Influence of Different Variable Selection Methods on Model Estimation Performance
Little previous research has investigated employing variable selection in machine learning-based remote sensing of vegetation chlorophyll contents. This research found that the optimal combination of variable selection methods and machine learning algorithms could produce more accurate SPAD estimation of wheat canopy. Among various combinations of three variable selection methods and four machine learning algorithms, the combination of RF and SVR_sigmoid demonstrates the best capability of SPAD estimation.
In this study, the RFR and SVR_linear models using the RFE variable selection method provided overall higher R
2 and more robust results than the RFR and SVR_linear models using RF or r variable selection methods. In addition, the number of optimal variables selected using RFE was much smaller than those selected using RF or r. Results from this study disagree with the superiority of RF over RFE as a variable selection method, as reported by Chen et al. [
27]. More research should be conducted to further evaluate these variable selection methods in agricultural remote sensing.
Using RF to select the optimal variables resulted in more accurate SPAD estimates compared with using the r variable selection method (
Table 6). However, the differences caused by using different variable selection methods are not as large as those caused by using different machine learning algorithms.
Three models produced the lowest RMSE and RRMSE (i.e., RFE-SVR_linear, RF-SVR_sigmoid, and r-SVR_sigmoid), as displayed in
Table 6. They had seven common optimal variables (i.e., green, VARI, GI, RGRI, RI, MCARI1, and MCARI) (
Table 5). This indicates that the seven variables are particularly important for accurate estimation of wheat canopy SPAD. The seven variables were selected from the 31 variables (
Table 2) that included 10 variables (i.e., NPCI, GCVI, CVI, MCARI1, MCARI, MTCI, NCARI, TCI, TCARI, and CIRE) proposed and widely used for monitoring chlorophyll or SPAD in previous studies [
16,
40,
54]. It is a bit surprising that, among the 10 variables, only two (i.e., MCARI1 and MCARI) are commonly selected by the three best SPAD estimation models in this study.
4.4. Limitations and Future Research
Although this preliminary study proposed a promising method for SPAD monitoring, there were still some limitations. This study found that the NDVI value of 0.25 could discriminate wheat from soil. When the new method is applied on other dates or on other farms, the threshold should be determined through a very careful visual check of the entire image.
Besides chlorophyll content, canopy reflectance is also sensitive to other influencing factors, such as canopy structure and leaf area index [
9,
15]. The error in the SPAD estimation of the RF-SVR_sigmoid model could be partially attributed to the omission of these influencing factors. Considering these influencing factors may further improve the SPAD estimation accuracies in future research.
This study applied 96 plots, and future research will employ a larger data base. In addition, this study involved 18 springness wheat varieties, and in contrast it involved only two weak springness varieties and four semi-winterness varieties. Hence, the wheat variety shares should be balanced in future research.
This study used only data from one growing season (i.e., the overwintering growth stage). Future research should collect data at more growth stages. In addition, this study involved only a single year, but variety screening often needs multiple-year experiments. Hence, the conclusions drawn in this study should be further evaluated in future studies with data from multiple growth stages and more years on various experimental farms.