Visual Growth Tracking for Automated Leaf Stage Monitoring Based on Image Sequence Analysis
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
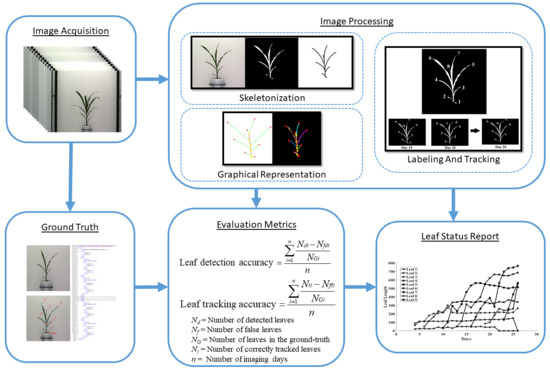
- View selection: Each plant is captured from multiple viewpoints to get a more accurate representation. We select the view at which the leaves are most distinct.
- Plant architecture determination: For each image in the sequence, we determine the architecture of the plant using a graph theoretic approach.
- Leaf tracking: The plant architectures are reconciled to determine the correspondences between the leaves over time to track them over the vegetative stage life cycle and demonstrate the temporal variation of the leaf-based phenotypes.
- Leaf status report: A leaf status report is produced as an output of the algorithm containing phenotypic information related to the entire life history of each leaf that best contributes to assessing plant vigor.
3.1. View Selection
3.2. Plant Architecture Determination
3.2.1. Segmentation
3.2.2. Skeletonization
- Base: The base of the plant is the point from which the stem of the plant emerges from the soil and is the lowest point of the skeleton.
- Collar/junction: The point at which a leaf is connected to the stem. The junctions, i.e., collars, are nodes of degree 3 or more in the graph.
- Tip: The free end of the leaf that is not connected to the stem.
- Leaf: The segments of the plant that connect the leaf tips and collars on the stem.
- Inter-junction: The segments of the plant connecting two collars are called inter-junctions.
| Algorithm 1 Plant architecture determination: produces a graphical representation of all the images of a plant sequence to detect the leaves and stem. |
| Input: The image sequence of a plant, i.e., } Output: G = {, , …, }, where is the graphical representation of the i-th image ∀i = 1,…, n. for do { // segment the image for day i // compute the skeleton of the segmented image . = // remove spurious edges with a threshold of pixels. // represent the plant skeleton as a graph // vertices and edges of the graph // degree 1 vertices // find the base ; // tips of the leaves // collars or junctions ; // Assign orientation to each leaf count = 1; // starting label for leaves for do { // follow junctions starting from base []=; // extract the vertices from edge if () continue; if () { // there is only one leaf at the junction .label = count; count ++; } else { // there are multiple leaves at the junction orient = count //previous leaf orientation orient = ¬ orient; // next leaf orientation leaves = // initialize with the leaves at the node while (l) do { // orientation of the next leaf // remove the leaf count ++; end while end for end for |
| Algorithm 2: determine the stem in a graph. |
| Input: A graph G and its base B. Output: A list of edges in G that constitute the stem (S). v = B; done = FALSE; S = []; while (¬ done) { = ; // find the non-visited adjacent vertex of degree ≠ 1 if ( = NULL) // there is no adjacent vertex with degree ≠1 done = TRUE; else{ S.append() // add the inter-stem segment to the stem ; } } |
3.3. Leaf Tracking
- A new leaf emerges above the last leaf in opposite alternate orientation, i.e., if the previous leaf emerged from the left side, the next leaf will emerge from the right side and will originate from a collar situated above the collar of the immediate previous leaf.
- In the event of a loss of a leaf, the height of its collar decreases, and the length of the corresponding inter-junction increases compared to the previous image.
- No more than one leaf may die in two consecutive images in a sequence.
- No more than one new leaf may emerge in two consecutive images in a sequence.
- 1
- Leaf emergence: A new leaf emerged, but no leaf was lost (Figure 5a).
- 2
- No change: No new leaf emerged, and no leaf was lost (Figure 5b). In this case, we transfer the labels from the previous graph to the next graph.
- 3
- Leaf loss: A leaf was lost, but no new leaf emerged (Figure 5c).
- 4
- Loss and emergence: A new leaf emerged, and a leaf was lost (Figure 5d).
| Algorithm 3 Leaf tracking algorithm. |
| Input: G = {, , …, , …, }, where is the graphical representation of the i-th image ∀i = 1,…, n, obtained from Algorithm 1. Output: = {, , …, , …, }, where is the graphical representation of the i-th image ∀i = 1,…, n, with the leaves correctly tracked and labeled. for do { lostLeaf = checkLostLeaf(, ); // See Equation (3) if (¬ lostLeaf) = firstLabel(); else = firstLabel() + 1; = updateGraph(, ); end for |
3.4. Leaf Status Report
Leaf Length
3.5. Evaluation Metrics
- Leaf-detection accuracy (LDA): The leaf-detection accuracy and leaf-tracking accuracy are respectively given by:where , , and are the number of detected leaves, the number of false leaves, and the actual number of leaves (as noted in the ground truth) for the i-th day for a given plant. This is computed for each plant separately.
- Leaf-tracking accuracy (LTA): This measures the accuracy of our leaf tracking algorithm and is given by:where , , and are the number of correctly tracked leaves, the number of wrongly tracked leaves, and the actual number of leaves (as noted in the ground truth) for the i-th day for a given plant, respectively. This is also computed for each plant separately.
4. UNL-CPPD
4.1. Imaging Setup
4.2. Dataset Organization
- Id: It serves two purposes based on its placement. If it is inside the Plant element, it serves the purpose of the image identifier, i.e., the day and the view of the image for which the information is represented in the XML document. When placed inside the Leaf element, it refers to the leaf number in order of emergence.
- Base: It has two attributes, i.e., x and y, which represent the pixel coordinates for the location of the base in the image.
- Leaf: It has four children, i.e., id, status, tip, and collar. id refers to the leaf emergence order, and status represents the status of the leaf (alive, dead, or missing). status “alive” means the leaf is visible in the image connected to a collar. status “dead” means that the leaf appears to be dead in the image mainly due to the separation from the stem. status “missing” means the leaf is no longer visible in the image either due to shedding or occlusion at a particular view. The tip element has children x and y, which represent the coordinates of the pixel location of the leaf tip; similarly, the collar element represents the coordinates of the pixel location of the junction.
5. Results
5.1. Leaf Status Report
5.2. Limitation Handling
- (1)
- Case A: The degrees of both the nodes are less than three. This case corresponds to the scenarios shown in Figure 10. In this case, the node with the most angular consistency with edge a is chosen. In Figure 11, Node 3 is selected, and edge b is marked with the current leaf number. When Node 2 is reached via edge c, the algorithm stops tracking since there are no unseen edges to follow, and it labels it as the tip for that leaf.
- (2)
- Case B: One node has degree three, and the other has degree two or less: This case may correspond to the scenarios in either Figure 11 or Figure 12. In this case, a two node look-ahead (from the new degree three node) is performed to identify all combinations of edges that form a path from Node 1 to the resulting nodes from the second look-ahead. The path with the highest angular consistency is chosen to continue the current leaf.Depending on the type of intersection, edge x is either shared (in the case of a crossover) or ignored (in the case of a tangential contact) for detecting leaves accurately. In Figure 10, the possible leaf segments are {}, and the path is selected for the highest angular consistency. When the algorithm reaches Node 5 from Node 3 following edge b, a similar analysis will continue the leaf to Node 6 using edge d, and edge x will remain unused and eventually ignored by the algorithm. For the scenario in Figure 12, however, with the same set of possible leaf segments, i.e., {}, the path is chosen for the highest angular consistency. When the algorithm reaches Node 2 from Node 4, the same analysis will select as the best path for leaf continuation, in essence sharing edge x. Thus, the leaves are detected accurately in the presence of different types of intersections.
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Xing, J.; Ai, H.; Liu, L.; Lao, S. Multiple Player Tracking in Sports Video: A Dual-Mode Two-Way Bayesian Inference Approach With Progressive Observation Modeling. IEEE Trans. Image Process. 2011, 20, 1652–1667. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Huynh, D.Q. Multiple Pedestrian Tracking From Monocular Videos in an Interacting Multiple Model Framework. IEEE Trans. Image Process. 2018, 27, 1361–1375. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Pupo, R.; Sierra-Romero, A.; Torres-Roman, D.; Shkvarko, Y.V.; Santiago-Paz, J.; Gómez-Gutiérrez, D.; Robles-Valdez, D.; Hermosillo-Reynoso, F.; Romero-Delgado, M. Vehicle Detection with Occlusion Handling, Tracking, and OC-SVM Classification: A High Performance Vision-Based System. Sensors 2018, 18, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, W.; Fan, M.; Guo, X.; Han, Q. A New Approach to Track Multiple Vehicles With the Combination of Robust Detection and Two Classifiers. IEEE Trans. Intell. Transp. Syst. 2018, 19, 174–186. [Google Scholar] [CrossRef]
- Yin, X.; Liu, X.; Chen, J.; Kramer, D.M. Joint Multi-Leaf Segmentation, Alignment, and Tracking from Fluorescence Plant Videos. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 1411–1423. [Google Scholar] [CrossRef] [Green Version]
- Cakir, R. Effect of water stress at different development stages on vegetative and reproductive growth of corn. Field Crop. Res. 2004, 89, 1–6. [Google Scholar] [CrossRef]
- Hardacre, A.K.; Turnbull, H.L. The Growth and Development of Maize (Zea mays L.) at Five Temperatures. Ann. Bot. 1986, 89, 779–787. [Google Scholar] [CrossRef]
- Choudhury, S.D.; Bashyam, S.; Qui, Y.; Samal, A.; Awada, T. Holistic and Component Plant Phenotyping using Visible Light Image Sequence. Plant Methods 2018, 19, 174–186. [Google Scholar] [CrossRef]
- Vylder, J.; Ochoa, D.; Philips, W.; Chaerle, L.; Straeten, D. Leaf Segmentation and Tracking using Probabilistic Parametric Active Contours. Vis. Comput. Graph. Collab. Tech. 2011, 6930, 75–85. [Google Scholar]
- Scharr, H.; Minervini, M.; French, A.P.; Klukas, C.; Kramer, D.M.; Liu, X.; Luengo, I.; Pape, J.; Polder, G.; Vukadinovic, D.; et al. Leaf segmentation in plant phenotyping: A collation study. Mach. Vis. Appl. 2016, 27, 585–606. [Google Scholar] [CrossRef] [Green Version]
- Jansen, M.; Gilmer, F.; Biskup, B.; Nagel, K.A.; Rascher, U.; Fischbach, A.; Briem, S.; Dreissen, G.; Tittmann, S.; Braun, S.; et al. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct. Plant Biol. 2009, 36, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Minervini, M.; Abdelsamea, M.M.; Tsaftaris, S.A. Image-based plant phenotyping with incremental learning and active contours. Ecol. Informatics 2014, 23, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Aksoy, E.E.; Abramov, A.; Wörgötter, F.; Scharr, H.; Fischbach, A.; Dellen, B. Modeling leaf growth of rosette plants using infrared stereo image sequences. Comput. Electron. Agric. 2015, 110, 78–90. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.D.; Goswami, S.; Bashyam, S.; Samal, A.; Awada, T. Automated Stem Angle Determination for Temporal Plant Phenotyping Analysis. In Proceedings of the ICCV workshop on Computer Vision Problmes in Plant Phenotyping, Venice, Italy, 28 October 2017; pp. 22–29. [Google Scholar]
- Maddonni, G.A.; Otegui, M.E.; Andrieu, B.; Chelle, M.; Casal, J.J. Maize leaves turn away from neighbors. Plant Physiol. 2002, 130, 1181–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassouna, M.S.; Farag, A.A. MultiStencils Fast Marching Methods: A Highly Accurate Solution to the Eikonal Equation on Cartesian Domains. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Latecki, L.J.; Liu, W.Y. Skeleton Pruning by Contour Partitioning with Discrete Curve Evolution. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ruberto, C.D. Recognition of shapes by attributed skeletal graphs. Pattern Recognit. 2004, 37, 21–31. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]












| Lost Leaf | New Leaf | Action |
|---|---|---|
| No | No | Transfer labels from to  |
| No | Yes | Transfer labels from to , and increment other labels  |
| Yes | No | Transfer labels from to ∀ ∈  |
| Yes | Yes | Transfer labels from to ∀ ∈ , and increment other labels  |
| Plant−ID | LDA | LTA | ||||
|---|---|---|---|---|---|---|
| 116 | 93 | 1 | 78 | 0.79 | 0.16 * | |
| 138 | 136 | 0 | 2 | 0.98 | 0.98 | |
| 142 | 140 | 0 | 8 | 0.98 | 0.94 | |
| 103 | 86 | 0 | 47 | 0.83 | 0.45 * | |
| 113 | 101 | 0 | 0 | 0.89 | 1.00 | |
| 122 | 120 | 3 | 2 | 0.96 | 0.98 | |
| 148 | 142 | 2 | 0 | 0.94 | 1.00 | |
| 149 | 138 | 0 | 0 | 0.93 | 1.00 | |
| 125 | 111 | 0 | 0 | 0.89 | 1.00 | |
| 141 | 131 | 0 | 0 | 0.93 | 1.00 | |
| 135 | 126 | 2 | 0 | 0.92 | 1.00 | |
| 144 | 140 | 0 | 0 | 0.97 | 1.00 | |
| 137 | 111 | 0 | 2 | 0.96 | 0.98 | |
| Average | 132 | 123 | <1 | 10.69 | 0.92 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashyam, S.; Choudhury, S.D.; Samal, A.; Awada, T. Visual Growth Tracking for Automated Leaf Stage Monitoring Based on Image Sequence Analysis. Remote Sens. 2021, 13, 961. https://doi.org/10.3390/rs13050961
Bashyam S, Choudhury SD, Samal A, Awada T. Visual Growth Tracking for Automated Leaf Stage Monitoring Based on Image Sequence Analysis. Remote Sensing. 2021; 13(5):961. https://doi.org/10.3390/rs13050961
Chicago/Turabian StyleBashyam, Srinidhi, Sruti Das Choudhury, Ashok Samal, and Tala Awada. 2021. "Visual Growth Tracking for Automated Leaf Stage Monitoring Based on Image Sequence Analysis" Remote Sensing 13, no. 5: 961. https://doi.org/10.3390/rs13050961
APA StyleBashyam, S., Choudhury, S. D., Samal, A., & Awada, T. (2021). Visual Growth Tracking for Automated Leaf Stage Monitoring Based on Image Sequence Analysis. Remote Sensing, 13(5), 961. https://doi.org/10.3390/rs13050961