Deep Learning and Phenology Enhance Large-Scale Tree Species Classification in Aerial Imagery during a Biosecurity Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ground Truth Data
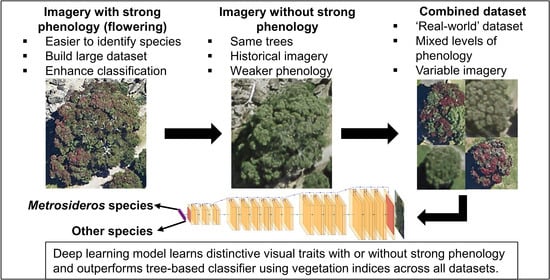
2.2. Imagery Datasets
2.3. Deep Learning Models
2.4. XGBoost Models
2.5. Performance Metrics
3. Results
3.1. XGBoost Models
3.2. Deep Learning Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldson, S.; Bourdôt, G.; Brockerhoff, E.; Byrom, A.; Clout, M.; McGlone, M.; Nelson, W.; Popay, A.; Suckling, D.; Templeton, M. New Zealand pest management: Current and future challenges. J. R. Soc. N. Z. 2015, 45, 31–58. [Google Scholar] [CrossRef]
- Kriticos, D.; Phillips, C.; Suckling, D. Improving border biosecurity: Potential economic benefits to New Zealand. N. Z. Plant Prot. 2005, 58, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kalaris, T.; Fieselmann, D.; Magarey, R.; Colunga-Garcia, M.; Roda, A.; Hardie, D.; Cogger, N.; Hammond, N.; Martin, P.T.; Whittle, P. The role of surveillance methods and technologies in plant biosecurity. In The Handbook of Plant Biosecurity; Springer: Dordrecht, The Netherlands, 2014; pp. 309–337. [Google Scholar]
- DiTomaso, J.M.; Van Steenwyk, R.A.; Nowierski, R.M.; Vollmer, J.L.; Lane, E.; Chilton, E.; Burch, P.L.; Cowan, P.E.; Zimmerman, K.; Dionigi, C.P. Enhancing the effectiveness of biological control programs of invasive species through a more comprehensive pest management approach. Pest. Manag. Sci. 2017, 73, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Keith, L.M.; Heller, W.P.; Hughes, M.A.; Vaughn, N.R.; Hughes, R.F.; Balzotti, C. A Spectral Mapping Signature for the Rapid Ohia Death (ROD) Pathogen in Hawaiian Forests. Remote Sens. 2018, 10, 404. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Asner, G.P. Applications of remote sensing to alien invasive plant studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Chen, G.; Potter, C.; Meentemeyer, R.K. Integrating multi-sensor remote sensing and species distribution modeling to map the spread of emerging forest disease and tree mortality. Remote Sens. Environ. 2019, 231, 111238. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Dungey, H.S. UAV Based Monitoring of Physiological Stress in Trees is Affected by Image Resolution and Choice of Spectral Index. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.O.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Krzystek, P.; Serebryanyk, A.; Schnörr, C.; Červenka, J.; Heurich, M. Large-scale mapping of tree species and dead trees in šumava national park and bavarian forest national park using lidar and multispectral imagery. Remote Sens. 2020, 12, 661. [Google Scholar] [CrossRef] [Green Version]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Dalponte, M.; Orka, H.O.; Gobakken, T.; Gianelle, D.; Naesset, E. Tree Species Classification in Boreal Forests With Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2012, 51, 2632–2645. [Google Scholar] [CrossRef]
- Hesketh, M.; Sánchez-Azofeifa, G.A. The effect of seasonal spectral variation on species classification in the Panamanian tropical forest. Remote Sens. Environ. 2012, 118, 73–82. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual tree crown segmentation and classification of 13 tree species using airborne hyperspectral data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef] [Green Version]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- De Lacerda, A.E.B.; Nimmo, E.R. Can we really manage tropical forests without knowing the species within? Getting back to the basics of forest management through taxonomy. For. Ecol. Manag. 2010, 259, 995–1002. [Google Scholar] [CrossRef]
- Van Horn, G.; Mac Aodha, O.; Song, Y.; Cui, Y.; Sun, C.; Shepard, A.; Adam, H.; Perona, P.; Belongie, S. The inaturalist species classification and detection dataset. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake, UT, USA, 18–23 June 2018; pp. 8769–8778. [Google Scholar]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef] [Green Version]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban tree species classification using a WorldView-2/3 and LiDAR data fusion approach and deep learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [Green Version]
- Mäyrä, J.; Keski-Saari, S.; Kivinen, S.; Tanhuanpää, T.; Hurskainen, P.; Kullberg, P.; Poikolainen, L.; Viinikka, A.; Tuominen, S.; Kumpula, T.; et al. Tree species classification from airborne hyperspectral and LiDAR data using 3D convolutional neural networks. Remote. Sens. Environ. 2021, 256, 112322. [Google Scholar] [CrossRef]
- Trier, Ø.D.; Salberg, A.-B.; Kermit, M.; Rudjord, Ø.; Gobakken, T.; Næsset, E.; Aarsten, D. Tree species classification in Norway from airborne hyperspectral and airborne laser scanning data. Eur. J. Remote. Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Cui, Y.; Song, Y.; Sun, C.; Howard, A.; Belongie, S. Large scale fine-grained categorization and domain-specific transfer learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake, UT, USA, 18–23 June 2018; pp. 4109–4118. [Google Scholar]
- Wäldchen, J.; Rzanny, M.; Seeland, M.; Mäder, P. Automated plant species identification—Trends and future directions. PLoS Comput. Biol. 2018, 14, e1005993. [Google Scholar] [CrossRef] [Green Version]
- Onishi, M.; Ise, T. Explainable identification and mapping of trees using UAV RGB image and deep learning. Sci. Rep. 2021, 11, 903. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Egli, S.; Höpke, M. CNN-Based Tree Species Classification Using High Resolution RGB Image Data from Automated UAV Observations. Remote Sens. 2020, 12, 3892. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.M.; Gloor, E.; Phillips, O.L.; Aragão, L.E.O.C. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef] [Green Version]
- Omer, G.; Mutanga, O.; Abdel-Rahman, E.M.; Adam, E. Performance of support vector machines and artificial neural network for mapping endangered tree species using WorldView-2 data in dukuduku forest, South Africa. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4825–4840. [Google Scholar] [CrossRef]
- Castro-Esau, K.L.; Sánchez-Azofeifa, G.A.; Rivard, B.; Wright, S.J.; Quesada, M. Variability in leaf optical properties of Mesoamerican trees and the potential for species classification. Am. J. Bot. 2006, 93, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, L.; Yin, D.; Li, X.; Diao, C.; Gong, H.; Shi, C.; Menenti, M.; Ge, Y.; Nie, S.; et al. Development of spectral-phenological features for deep learning to understand Spartina alterniflora invasion. Remote Sens. Environ. 2020, 242, 111745. [Google Scholar] [CrossRef]
- Carnegie, A.J.; Kathuria, A.; Pegg, G.S.; Entwistle, P.; Nagel, M.; Giblin, F.R. Impact of the invasive rust Puccinia psidii (myrtle rust) on native Myrtaceae in natural ecosystems in Australia. Biol. Invasions 2016, 18, 127–144. [Google Scholar] [CrossRef] [Green Version]
- Glen, M.; Alfenas, A.C.; Zauza, E.A.V.; Wingfield, M.J.; Mohammed, C. Puccinia psidii: A threat to the Australian environment and economy—A review. Australas. Plant Pathol. 2007, 36, 1–16. [Google Scholar] [CrossRef]
- Carnegie, A.J.; Cooper, K. Emergency response to the incursion of an exotic myrtaceous rust in Australia. Australas. Plant Pathol. 2001, 40, 346. [Google Scholar] [CrossRef]
- Coutinho, T.A.; Wingfield, M.J.; Alfenas, A.C.; Crous, P.W. Eucalyptus Rust: A Disease with the Potential for Serious International Implications. Plant Dis. 1998, 82, 819–825. [Google Scholar] [CrossRef] [Green Version]
- McTaggart, A.R.; Roux, J.; Granados, G.M.; Gafur, A.; Tarrigan, M.; Santhakumar, P.; Wingfield, M.J. Rust (Puccinia psidii) recorded in Indonesia poses a threat to forests and forestry in South-East Asia. Australas. Plant Pathol. 2015, 45, 83–89. [Google Scholar] [CrossRef]
- Roux, J.; Greyling, I.; Coutinho, T.A.; Verleur, M.; Wingfield, M.J. The Myrtle rust pathogen, Puccinia psidii, discovered in Africa. IMA Fungus 2013, 4, 155–159. [Google Scholar] [CrossRef]
- De Lange, P.J.; Rolfe, J.R.; Barkla, J.W.; Courtney, S.P.; Champion, P.D.; Perrie, L.R.; Beadel, S.M.; Ford, K.A.; Breitwieser, I.; Schoenberger, I.; et al. Conservation Status of New Zealand Indigenous Vascular Plants, 2017; Department of Conservation: Wellington, New Zealand, 2018; ISBN 978-1-98-85146147-1.
- Allan, H.H. Flora of New Zealand Volume I Indigenous Tracheophyta-Psilopsida, Lycopsida, Filicopsida, Gymnospermae, Dicotyledones; Flora of New Zealand-Manaaki Whenua Online Reprint Series; Government Printer Publication: Wellington, New Zealand, 1982; Volume 1, ISBN 0-477-01056-3.
- Loope, L. A summary of information on the rust Puccinia psidii Winter (guava rust) with emphasis on means to prevent introduction of additional strains to Hawaii. In Open-File Report; US Geological Survey: Reston, VA, USA, 2010; pp. 1–31. Available online: https://pubs.usgs.gov/of/2010/1082/of2010-1082.pdf (accessed on 17 June 2019).
- Sandhu, K.S.; Park, R.F. Genetic Basis of Pathogenicity in Uredo Rangelii; University of Sydney: Camperdown, Sydney, 2013. [Google Scholar]
- Ho, W.H.; Baskarathevan, J.; Griffin, R.L.; Quinn, B.D.; Alexander, B.J.R.; Havell, D.; Ward, N.A.; Pathan, A.K. First Report of Myrtle Rust Caused by Austropuccinia psidii on Metrosideros kermadecensis on Raoul Island and on M. excelsa in Kerikeri, New Zealand. Plant Dis. 2019, 103, 2128. [Google Scholar] [CrossRef]
- Beresford, R.M.; Turner, R.; Tait, A.; Paul, V.; Macara, G.; Yu, Z.D.; Lima, L.; Martin, R. Predicting the climatic risk of myrtle rust during its first year in New Zealand. N. Z. Plant Prot. 2018, 71, 332–347. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Zvoleff, A. Glcm: Calculate Textures from Grey-Level Co-Occurrence Matrices (GLCMs). R-CRAN Project. 2019. Available online: https://cran.r-project.org/web/packages/glcm/index.html (accessed on 14 June 2019).
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Hall-Beyer, M. Practical guidelines for choosing GLCM textures to use in landscape classification tasks over a range of moderate spatial scales. Int. J. Remote Sens. 2017, 38, 1312–1338. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme gradient boosting. R Package Version 2019, 1, 0.4–2. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Pham, L.T.; Brabyn, L.; Ashraf, S. Combining QuickBird, LiDAR, and GIS topography indices to identify a single native tree species in a complex landscape using an object-based classification approach. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 187–197. [Google Scholar] [CrossRef]
- Dymond, C.C.; Mladenoff, D.J.; Radeloff, V.C. Phenological differences in Tasseled Cap indices improve deciduous forest classification. Remote Sens. Environ. 2002, 80, 460–472. [Google Scholar] [CrossRef]
- Wolter, P.T.; Mladenoff, D.J.; Host, G.E.; Crow, T.R. Improved forest classification in the Northern Lake States using multi-temporal Landsat imagery. Photogramm. Eng. Remote Sens. 1995, 61, 1129–1144. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zörner, J.; Dymond, J.R.; Shepherd, J.D.; Wiser, S.K.; Jolly, B. LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand. Forests 2018, 9, 702. [Google Scholar] [CrossRef] [Green Version]
- MacFaden, S.W.; O’Neil-Dunne, J.P.; Royar, A.R.; Lu, J.W.; Rundle, A.G. High-resolution tree canopy mapping for New York City using LIDAR and object-based image analysis. J. Appl. Remote Sens. 2012, 6, 063567. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef]








| Imagery Dataset | Phenology | Resolution, Colour Channels |
|---|---|---|
| Tauranga—summer 2018–2019 | Wide-spread flowering | 10 cm/pixel, 3-band RGB |
| Tauranga—March 2017 | Limited flowering | 10 cm/pixel, 3-band RGB |
| Dataset | Purpose | Tree Counts (Pōhutukawa/Other spp.) | Data Splits Training/Validation/Test |
|---|---|---|---|
| Tauranga 2019 | Classification using phenology | 2300 (1150/1150) | 1610/345/345 (70/15/15%) |
| Tauranga 2017 | Classification without phenology | 2300 (1150/1150) | 1610/345/345 (70/15/15%) |
| Tauranga 2017 and 2019 combined | Combined classification with and without phenology | 4600 (2300/2300) | 3220/690/690 (70/15/15%) |
| Variable Name | Description | Definition | Source |
|---|---|---|---|
| Mean red | Mean of red channel DNs | NA | |
| Mean green | Mean of green channel DNs | NA | |
| Mean blue | Mean of blue channel DNs | NA | |
| SD red | Standard deviation of red channel DNs | NA | |
| SD green | Standard deviation of green channel DNs | NA | |
| SD blue | Standard deviation of blue channel DNs | NA | |
| RG ratio | Red green ratio index | [61] | |
| Normdiff RG | Normalised difference red/green ratio | NA | |
| Scaled red | Scaled red ratio | NA | |
| Scaled green (SG) | Scaled green ratio | NA | |
| Scaled blue | Scaled blue ratio | NA | |
| SD GI | Standard deviation of the scaled green index | NA | |
| GLCM correlation | Textural metric computed on RGB channels | Grey-level co-occurrence correlation | [55] |
| GLCM homogeneity | Textural metric computed on RGB channels | Grey-level co-occurrence homogeneity | [55] |
| GLCM mean | Textural metric computed on RGB channels | Grey-level co-occurrence mean | [55] |
| GLCM entropy | Textural metric computed on RGB channels | Grey-level co-occurrence entropy | [55] |
| Metric | Description | Definition |
|---|---|---|
| Accuracy | A measure of how often the classifier’s predictions were correct. | |
| Error | A measure of how often the classifier’s predictions were wrong. | |
| Cohen’s kappa | A measure of a classifier’s prediction accuracy that accounts for chance agreement. | |
| Precision (Positive predictive value) | A measure of the proportion of positive predictions that were correct. | |
| Sensitivity (Recall) | The proportion of actual positives (Metrosideros) that were correctly identified by the classifier. | |
| Specificity | The proportion of actual negatives (other species) that were correctly identified by the classifier. |
| Classification with Strong Phenology (2019) | Classification without Strong Phenology (2017) | Classification of Combined 2017 & 2019 Datasets with and without Phenology | |
|---|---|---|---|
| XGBoost | |||
| Accuracy | 86.7% | 79.4% | 83.2% |
| Error | 13.3% | 20.6% | 16.8% |
| kappa | 0.733 | 0.588 | 0.664 |
| Precision (PPV) | 0.861 | 0.793 | 0.831 |
| Sensitivity (recall) | 0.871 | 0.788 | 0.827 |
| Specificity | 0.863 | 0.800 | 0.837 |
| Deep Learning | |||
| Accuracy | 97.4% | 92.7% | 95.2% |
| Error | 3.6% | 7.3% | 4.8% |
| kappa | 0.948 | 0.855 | 0.904 |
| Precision (PPV) | 0.982 | 0.939 | 0.973 |
| Sensitivity (recall) | 0.965 | 0.912 | 0.932 |
| Specificity | 0.983 | 0.943 | 0.973 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pearse, G.D.; Watt, M.S.; Soewarto, J.; Tan, A.Y.S. Deep Learning and Phenology Enhance Large-Scale Tree Species Classification in Aerial Imagery during a Biosecurity Response. Remote Sens. 2021, 13, 1789. https://doi.org/10.3390/rs13091789
Pearse GD, Watt MS, Soewarto J, Tan AYS. Deep Learning and Phenology Enhance Large-Scale Tree Species Classification in Aerial Imagery during a Biosecurity Response. Remote Sensing. 2021; 13(9):1789. https://doi.org/10.3390/rs13091789
Chicago/Turabian StylePearse, Grant D., Michael S. Watt, Julia Soewarto, and Alan Y. S. Tan. 2021. "Deep Learning and Phenology Enhance Large-Scale Tree Species Classification in Aerial Imagery during a Biosecurity Response" Remote Sensing 13, no. 9: 1789. https://doi.org/10.3390/rs13091789
APA StylePearse, G. D., Watt, M. S., Soewarto, J., & Tan, A. Y. S. (2021). Deep Learning and Phenology Enhance Large-Scale Tree Species Classification in Aerial Imagery during a Biosecurity Response. Remote Sensing, 13(9), 1789. https://doi.org/10.3390/rs13091789