How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Methods
2.2.1. Data Resources and Preprocessing
2.2.2. Collection of Forest Inventory Data
2.2.3. Calculation of Forest Phenological Metrics
2.2.4. Classification and Accuracy Assessment
3. Results
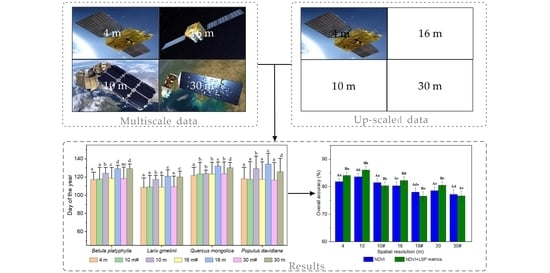
3.1. Multiscale Sequence NDVI Curve of Different Deciduous Forest Stands
3.2. Multiscale SOS Results by Satellite Images
3.3. Multiscale Classification by Satellite Images and Up-Scaled Images
3.4. Contributions of Different LSP Metrics to Multiscale Tree Species Classification
3.5. Comparisons of Accuracy for Producer and User of Dominant Tree Species
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Month | Imaging Date (Gaofen-2) | Imaging Date (Sentinel-2) | Imaging Date (Gaofen-1) | Imaging Date (Landsat-8) |
|---|---|---|---|---|
| January | 20180121 | 20180120 | 20180117 | 20180111 |
| February | 20180215 | 20180214 | 20180214 | 20180212 |
| March | 20180316 | 20180311 | 20180311 | 20180316 |
| April | 20180430 | 20180415 | 20180418 | 20180417 |
| May | 20180510 | 20180515 | 20180515 | 20180519 |
| June | 20180608 | 20180614 | 20180621 | 20180620 |
| July | 20180722 | 20180729 | 20180722 | 20180706 |
| August | 20180816 | 20180823 | 20180820 | 20180823 |
| September | 20170920 | 20170922 | 20180916 | 20180924 |
| October | 20181019 | 20181017 | 20181019 | 20181025 |
| November | 20181118 | 20181121 | 20181120 | 20181126 |
| December | 20181213 | 20181216 | 20181215 | 20181213 |
| Species Name | Picture | Main Characteristics |
|---|---|---|
| Larix gmelinii (Lg) |  | A deciduous tree (30 m), D.B.H. 90 cm or so. The barks are taupe, and the branches consist of obvious long branches and short branches. Light-loving, cold-resistant, drought-resistant. Mainly distributed in Northeast China as well as the mountainous areas from eastern Inner Mongolia to eastern Siberia of Russia. |
| Populus davidiana (Pd) |  | A deciduous tree (25 m), D.B.H. 60 cm or so. The barks are gray or gray-green, and the leaves are ovate or nearly round with incised margin. Light-loving, cold-resistant, and soil are adaptable. Mainly distributed in valleys of high mountains in Northeast China, Northwest China, North China and Southwest China. |
| Betula platyphylla (Bp) |  | A deciduous tree (27 m), D.B.H. 80 cm or so. The barks are white and smooth or local cracking, and the leaves are ovate with a jagged margin. Light-loving, cold-loving, and soil adaptable. Mainly distributed in Northeast China, North China and the mountainous areas on the outer edge of the Qinghai-Tibet Plateau. |
| Quercus mongolica (Qm) |  | A deciduous tree (30 m), D.B.H. 60 cm or so. The barks are dark taupe, the branches are purplish-brown, the leaves are obovate with around margin. Light-loving, soil adaptable. Mainly distributed in Northeast China and part of northern North China. |
References
- Sannier, C.; McRoberts, R.E.; Fichet, L. Suitability of global forest change data to report forest cover estimates at national level in Gabon. Remote Sens. Environ. 2016, 173, 326–338. [Google Scholar] [CrossRef]
- Feng, Y.; Lu, D.S.; Chen, Q.; Keller, M.; Moran, E.; Dos-Santo, M.N.; Bolf, E.L.; Batistella, M. Examining effective use of data sources and modeling algorithms for improving biomass estimation in a moist tropical forest of the Brazilian Amazon. Int. J. Digit. Earth 2017, 10, 996–1016. [Google Scholar] [CrossRef]
- Frenne, P.D.; Zellweger, F.; Rodríguez-Sánchez, F.; Scheffers, B.R.; Hylander, K.; Luoto, M.; Vellend, M.; Verheyen, K.; Lenoir, J. Global buffering of temperatures under forest canopies. Nat. Ecol. Evol. 2019, 3, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Heinzel, J.; Koch, B. Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int. J. Appl. Earth Obs. 2012, 18, 101–110. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Meddens, A.H.; Hicke, J.A.; Vierling, L.A. Evaluating the potential of multispectral imagery to map multiple stages of tree mortality. Remote Sens. 2011, 115, 1632–1642. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Tang, L.N.; Shao, G.F. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Immitzer, M.; Böck, S.; Einzmann, K.; Vuolo, F.; Pinnel, N.; Wallner, A.; Atzberger, C. Fractional cover mapping of spruce and pine at 1 ha resolution combining very high and medium spatial resolution satellite imagery. Remote Sens. Environ. 2017, 204, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Li, Z.L. Scale issues in remote sensing: A review on analysis, processing and modeling. Sensors 2009, 9, 1768–1793. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.L.; Liu, D.S. Accurate mapping of forest types using dense seasonal Landsat time-series. ISPRS J. Photogramm. Remote Sens. 2014, 96, 1–11. [Google Scholar] [CrossRef]
- Roth, K.l; Roberts, D.A.; Dennison, P.E.; Peterson, S.H.; Alonzo, M. The impact of spatial resolution on the classification of plant species and functional types within imaging spectrometer data. Remote Sens. Environ. 2015, 171, 45–57. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S.; Yu, Q. Assessing the potential of multi-seasonal high resolution Pléiades satellite imagery for mapping urban tree species. Int. J. Appl. Earth Obs. Geoinf. 2018, 71, 144–158. [Google Scholar] [CrossRef]
- Dudley, K.L.; Dennison, P.E.; Roth, K.L.; Roberts, D.A.; Coates, A.R. A multi-temporal spectral library approach for mapping vegetation species across spatial and temporal phenological gradients. Remote Sens. Environ. 2015, 167, 121–134. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest stand species mapping using the Sentinel-2 time series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef] [Green Version]
- Kempeneers, P.; Sedano, F.; Seebach, L.M.; Strobl, P.; San-Miguel-Ayanz, J. Data fusion of different spatial resolution remote sensing images applied to forest-type mapping. IEEE T. Geocsi. Remote. 2011, 49, 4977–4986. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Granero-Belinchon, C.; Adeline, K.; Briottet, X. Impact of the number of dates and their sampling on a NDVI time series reconstruction methodology to monitor urban trees with Venμs satellite. Int. J. Appl. Earth Obs. Geoinf. 2021, 95, 102257. [Google Scholar] [CrossRef]
- Xu, K.J.; Tian, Q.J.; Zhang, Z.Y.; Yue, J.B.; Chang, C.T. Tree species (genera) identification with GF-1 time-series in a forested landscape, Northeast China. Remote Sens. 2020, 12, 1554. [Google Scholar] [CrossRef]
- Kollert, A.; Bremer, M.; Löw, M.; Rutzinger, M. Exploring the potential of land surface phenology and seasonal cloud free composites of one year of Sentinel-2 imagery for tree species mapping in a mountainous region. Int. J. Appl. Earth Obs. 2021, 94, 102208. [Google Scholar] [CrossRef]
- Li, X.; Chen, W.Y.; Sanesi, G.; Lafortezza, R. Remote sensing in urban forestry: Recent applications and future directions. Remote Sens. 2019, 11, 1144. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.A.; Wilson, A.K.; George, M.; Hinsley, S.A. Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Appl. Veg. Sci. 2010, 13, 86–99. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Wei, X.; Yao, Y.; Su, Y.; Jiang, B.; Wang, X. Land cover classification of Landsat data with phenological features extracted from time series MODIS NDVI data. Remote Sens. 2014, 6, 11518–11532. [Google Scholar] [CrossRef] [Green Version]
- Masemola, C.; Cho, M.A.; Ramoelo, A. Sentinel-2 time series based optimal features and time window for mapping invasive Australian native Acacia species in KwaZulu Natal, South Africa. Int. J. Appl. Earth Obs. Geoinf. 2020, 93, 102207. [Google Scholar] [CrossRef]
- Gessner, U.; Machwitz, M.; Conrad, C.; Dechab, S. Estimating the fractional cover of growth forms and bare surface in savannas. A multi-resolution approach based on regression tree ensembles. Remote Sens. Environ. 2013, 129, 90–102. [Google Scholar] [CrossRef] [Green Version]
- Achard, F.; Estreguil, C. Forest classification of Southeast Asia using NOAA AVHRR data. Remote Sens. Environ. 1995, 54, 198–208. [Google Scholar] [CrossRef]
- Xiao, X.M.; Boles, S.; Liu, J.Y.; Zhuang, D.F.; Liu, M.L. Characterization of forest types in Northeastern China, using multi-temporal SPOT-4 VEGETATION sensor data. Remote Sens. Environ. 2002, 82, 335–348. [Google Scholar] [CrossRef]
- Yu, X.F.; Zhuang, D.F.; Chen, H.; Hou, X.Y. Forest classification based on MODIS time series and vegetation phenology. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Anchorage, AK, USA, 20–24 September 2004; Volume 4, pp. 2369–2372. [Google Scholar] [CrossRef]
- Pimple, U.; Sitthi, A.; Simonetti, D.; Pungkul, S.; Leadprathom, K.; Chidthaisong, A. Topographic correction of Landsat TM-5 and Landsat OLI-8 imagery to improve the performance of forest classification in the mountainous terrain of Northeast Thailand. Sustainability 2017, 9, 258. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Khamzina, A.; Pflugmacher, D.; Martius, C. Forest cover mapping in post-Soviet Central Asia using multi-resolution remote sensing imagery. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Townshend, J.R.; Masek, J.G.; Huang, C.; Vermote, E.F.; Gao, F.; Channan, S.; Sexton, J.O.; Feng, M.; Narasimhan, R.; Kim, D.; et al. Global characterization and monitoring of forest cover using Landsat data: Opportunities and challenges. Int. J. Digit. Earth 2012, 5, 373–397. [Google Scholar] [CrossRef] [Green Version]
- Ota, T.; Mizoue, N.; Yoshida, S. Influence of using texture information in remote sensed data on the accuracy of forest type classification at different levels of spatial resolution. J. For. Res. 2011, 16, 432–437. [Google Scholar] [CrossRef]
- Pu, R.L.; Bell, S. Mapping seagrass coverage and spatial patterns with high spatial resolution IKONOS imagery. Int. J. Appl. Earth Obs. Geoinf. 2017, 54, 145–158. [Google Scholar] [CrossRef]
- Puzzolo, V.; Denatale, F.; Gianne, F. Forest species discrimination in an Alpine mountain area using a fuzzy classification of multi-temporal SPOT (HRV) data. IEEE Int. Geosci. Remote Sens. Symp. 2003, 4, 2538–2540. [Google Scholar] [CrossRef]
- Persson, M.; Lindberg, E.; Reese, H. Tree species classification with multi-temporal Sentinel-2 data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef] [Green Version]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of different machine learning algorithms for scalable classification of tree types and tree species based on Sentinel-2 data. Remote Sens. 2018, 10, 1419. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; White, J.C.; Wulder, M.A. Optical remotely sensed time series data for land cover classification: A review. ISPRS J. Photogramm. Remote Sens. 2016, 116, 55–72. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.H.; Pan, Y.Z.; Zhu, X.F.; Zhu, W.Q. Using phenological metrics and the multiple classifier fusion method to map land cover types. J. Appl. Remote Sens. 2014, 8, 083691. [Google Scholar] [CrossRef]
- Michez, A.; Piegay, H.; Jonathan, L.; Claessens, H.; Lejeune, P. Mapping of riparian invasive species with supervised classification of Unmanned Aerial System (UAS) imagery. Int. J. Appl. Earth Obs. 2015, 44, 88–94. [Google Scholar] [CrossRef]
- Xu, K.J.; Tian, Q.J.; Xu, N.X.; Yue, J.B.; Tang, S.F. Classifying forest dominant trees species based on high dimensional time-series NDVI data and differential transform methods. Spectrosc. Spectr. Anal. 2019, 39, 3794–3800. [Google Scholar] [CrossRef]
- Kong, J.X.; Zhang, Z.C.; Zhang, J. Classification and identification of plant species based on multi-source remote sensing data: Research progress and prospect. Biodivers. Sci. 2019, 27, 796–812. [Google Scholar] [CrossRef]
- Peng, D.; Wu, C.; Zhang, X.; Yu, L.; Huete, A.R.; Wang, F.; Luo, S.; Liu, X.; Zhang, H. Scaling up spring phenology derived from remote sensing images. Agric. For. Meteorol. 2018, 256–257, 207–219. [Google Scholar] [CrossRef]
- Zeng, L.L.; Wardlow, B.D.; Xiang, D.X.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Peng, D.; Zhang, X.; Zhang, B.; Liu, L.; Liu, X.; Huete, A.R.; Huang, W.; Wang, S.; Luo, S.; Zhang, X.; et al. Scaling effects on spring phenology detections from MODIS data at multiple spatial resolutions over the contiguous United States. ISPRS J. Photogramm. Remote Sens. 2017, 132, 185–198. [Google Scholar] [CrossRef]
- Ge, Q.; Dai, J.; Cui, H.; Wang, H. Spatiotemporal variability in start and end of growing season in China related to climate variability. Remote Sens. 2016, 8, 433. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Cao, R.; Shen, M.; Chen, J.; Zhang, X. How does scale effect influence spring vegetation phenology estimated from satellite-derived vegetation indexes? Remote Sens. 2019, 11, 2137. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.J.; Zeng, Y.N.; Zhang, M. Mapping rice paddy distribution by using time series HJ blend data and phenological parameters. J. Remote Sens. 2018, 22, 381–391. [Google Scholar] [CrossRef]
- Schwieder, M.; Leitão, P.J.; Pinto, J.R.; Teixeira, A.C.; Pedroni, F.; Sanchez, M.; Bustamante, M.M.; Hostert, P. Landsat phenological metrics and their relation to aboveground carbon in the Brazilian Savanna. Carbon Balanc. Manag. 2018, 13, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Schaaf, A.; Dennison, P.; Fryer, G.; Roth, K.; Roberts, D. Mapping plant functional types at multiple spatial resolutions using imaging spectrometer data. GISci. Remote Sens. 2011, 48, 324–344. [Google Scholar] [CrossRef]
- Peña, M.A.; Cruz, P.; Roig, M. The effect of spectral and spatial degradation of hyperspectral imagery for the sclerophyll tree species classification. Int. J. Remote Sens. 2013, 34, 7113–7130. [Google Scholar] [CrossRef]
- Xu, K.J.; Tian, Q.J.; Yang, Y.J.; Yue, J.B.; Tang, S.F. How up-scaling of remote-sensing images affects land-cover classification by comparison with multiscale satellite images. Int. J. Remote Sens. 2019, 40, 2784–2810. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Gao, F.; Liu, Y.; Schaaf, C.; Friedl, M.; Yu, Y.; Jayavelu, S.; Gray, J.; Liu, L.; et al. Exploration of scaling effects on coarse resolution land surface phenology. Remote Sens. Environ. 2017, 190, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.Q.; Zhu, X.L.; Wu, J.; Shen, M.G.; Chen, J. Coarse-resolution satellite images overestimate urbanization effects on vegetation spring phenology. Remote Sens. 2020, 12, 117. [Google Scholar] [CrossRef] [Green Version]
- Moody, A.; Woodcock, C.E. Scale-dependent errors in the estimation of land-cover proportions: Implications for global land-cover datasets. Photogramm. Eng. Remote Sens. 1994, 60, 585–594. [Google Scholar]
- Luan, H.J.; Tian, Q.J.; Yu, T.; Hu, X.L.; Huang, Y.; Liu, L.; Du, L.T.; Wei, X. Review of up-scaling of quantitative remote sensing. Adv. Earth Sci. 2013, 28, 657–664. [Google Scholar] [CrossRef]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky-Golay filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Wang, Y.F.; Xue, Z.H.; Chen, J.; Chen, G.Z. Spatio-temporal analysis of phenology in Yangtze river delta based on MODIS NDVI time series from 2001 to 2015. Front. Earth Sci. 2019, 13, 92–110. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R.S.; Wheeler, B.; Iiames, J.S.; Campbell, J.B. An evaluation of time-series smoothing algorithms for land-cover classifications using MODIS-NDVI multi-temporal data. Remote Sens. Environ. 2016, 174, 258–265. [Google Scholar] [CrossRef]
- Xu, K.J.; Zeng, H.D.; Zhu, X.B.; Tian, Q.J. Evaluation of five commonly used atmospheric correction algorithms for multi-temporal aboveground forest carbon storage estimation. Spectrosc. Spect. Anal. 2017, 37, 3493–3498. [Google Scholar] [CrossRef]
- Jönnson, P.; Eklundh, L. TIMESAT-a program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.T.; Wang, H.C.; Huang, C. Impact of vegetation onset time on the net primary productivity in a mountainous island in Pacific Asia. Environ. Res. Lett. 2013, 8, 05030. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.T.; Guan, H.D.; Shen, M.G.; Liang, W.; Jiang, L. Changes in autumn vegetation dormancy onset date and the climate controls across temperate ecosystems in China from 1982 to 2010. Glob. Chang. Biol. 2014, 21, 1–14. [Google Scholar] [CrossRef]
- Heumann, B.W.; Seaquist, J.W.; Eklundh, L.; Jönsson, P. AVHRR derived phenological change in the Sahel and Soudan, Africa, 1982-2005. Remote Sens. Environ. 2007, 108, 385–392. [Google Scholar] [CrossRef]
- Jiao, F.S.; Liu, H.Y.; Xu, X.J.; Gong, H.B.; Lin, Z.S. Trend evolution of vegetation phenology in China during the period of 1981–2016. Remote Sens. 2020, 12, 572. [Google Scholar] [CrossRef] [Green Version]
- Lebrini, Y.; Boudhar, A.; Hadria, R.; Lionboui, H.; Elmansouri, L.; Arrach, R.; Ceccato, P.; Benabdelouahab, T. Identifying agricultural systems using SVM classification approach based on phenological metrics in a semi-arid region of Morocco. Earth Syst. Environ. 2019, 3, 277–288. [Google Scholar] [CrossRef]
- Sothe, C.; de Almeida, C.M.; Liesenberg, V.; Schimalski, M.B. Evaluating Sentinel-2 and Landsat-8 data to map sucessional forest stages in a subtropical forest in southern Brazil. Remote Sens. 2017, 9, 838. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First experience with Sentinel-2 data for crop and tree species classifications in central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăgut, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with random forest using very high spatial resolution 8-band WorldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Palomeque, G.; Dupuy, J.M.; Portillo-Quintero, C.A.; Andrade, J.L.; Tun-Dzul, F.J.; Hernández-Stefanoni, J.L. Mapping forest age and characterizing vegetation structure and species composition in tropical dry forests. Ecol. Indic. 2021, 120, 106955. [Google Scholar] [CrossRef]
- Chutia, D.; Bhattacharyya, D.K.; Sarma, K.K.; Kalita, R.; Sudhakar, S. Hyperspectral remote sensing classifications: A perspective survey. Trans. GIS 2016, 20, 463–490. [Google Scholar] [CrossRef]
- Shang, X.; Chisholm, L. Classification of Australian native forest species using hyperspectral remote sensing and machine-learning classification algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2481–2489. [Google Scholar] [CrossRef]
- Han, H.; Lee, S.; Im, J.; Kim, M.; Lee, M.I.; Ahn, M.; Chung, S.R. Detection of convective initiation using meteorological imager onboard communication, ocean, and meteorological satellite based on machine learning approaches. Remote Sens. 2015, 7, 9184–9204. [Google Scholar] [CrossRef] [Green Version]
- Immitzer, M.; Neuwirth, M.; Bck, S.; Brenner, H.; Atzberger, C. Optimal input features for tree epecies classification in central Europe based on multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef] [Green Version]
- Janssen, L.L.F.; van der Wel, F.J.M. Accuracy assessment of satellite derived land-gover data: A review. Photogramm. Eng. Remote Sens. 1994, 60, 419–426. [Google Scholar] [CrossRef]
- Yang, Y.; Xiao, P.; Feng, X.; Li, H. Accuracy assessment of seven global land cover datasets over China. ISPRS J. Photogramm. Remote Sens. 2017, 125, 156–173. [Google Scholar] [CrossRef]
- Richter, R.; Reu, B.; Wirth, C.; Doktor, D.; Vohland, M. The use of airborne hyperspectral data for tree species classification in a species-rich Central European forest area. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 464–474. [Google Scholar] [CrossRef]
- Wang, L.; Yang, R.; Tian, Q.; Yang, Y.; Zhou, Y.; Sun, Y.; Mi, X. Comparative analysis of GF-1 WFV, ZY-3 MUX, and HJ-1 CCD sensor data for grassland monitoring applications. Remote Sens. 2015, 7, 2089–2108. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Xie, G.D.; Zhou, D.M.; Zhang, C.S.; Jiao, C.C. Remote sensing classification of marsh wetland with different resolution images. J. Resour. Ecol. 2016, 7, 107–114. [Google Scholar] [CrossRef]
- Lessel, J.; Ceccato, P. Creating a basic customizable framework for crop detection using Landsat imagery. Int J. Remote Sens. 2016, 37, 6097–6107. [Google Scholar] [CrossRef]
- Hu, Y.F.; Xu, Z.Y.; Liu, Y.; Yan, Y.; Wang, Q.Q. Accuracy analysis of up-scaling data: A case study with land use data in Xilin Gol of Inner Mongolia, China. Geogr. Res. 2012, 31, 1961–1972. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, Y.U.; Meng, Q.; Xinli, H.U.; Chang, L.I. Image Quality Evaluation of Multi-Scale Resampling in Geometric Correction. J. Huazhong Norm. Univ. 2013, 47, 426–430. [Google Scholar] [CrossRef]
- Melaas, E.K.; Friedl, M.A.; Zhu, Z. Detecting interannual variation in deciduous broadleaf forest phenology using Landsat TM/ETM+ data. Remote Sens. Environ. 2013, 132, 176–185. [Google Scholar] [CrossRef]
- Tang, S.F.; Tian, Q.J.; Xu, K.J.; Xu, N.X.; Yue, J.B. Age information retrieval of Larix gmelinii forest using Sentinel-2 data. J. Remote Sens. 2020, 24, 1511–1524. [Google Scholar] [CrossRef]
- Hay, G.J.; Niernann, K.O.; Goodenough, D.J. Spatial thresholds, image-objects, and upscaling: A multiscale evaluation. Remote Sens. Environ. 1997, 62, 1–19. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, Y.; Zhang, Y.; Knyazikhin, Y.; Bogaert, J.; Mynemi, R.B. Radiative transfer based scaling of LAI retrieval from reflectance data of different resolutions. Remote Sens. Environ. 2002, 84, 143–159. [Google Scholar] [CrossRef]
- Wu, H.; Tang, B.H.; Li, Z.L. Impact of nonlinearity and discontinuity on the spatial scaling effects of the leaf area index retrieved from remotely sensed data. Int. J. Remote Sens. 2013, 34, 3503–3519. [Google Scholar] [CrossRef]
- Jiang, J.; Xiao, Z.; Wang, J.; Song, J. Multiscale estimation of leaf area index from satellite observations based on an ensemble multiscale filter. Remote Sens. 2016, 8, 229. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.Q.; Huang, W.J.; Niu, Z.; Wang, C.Y.; Li, W.; Yu, B. Validation of synthetic daily Landsat NDVI time series data generated by the improved spatial and temporal data fusion approach. Inf. Fusion 2018, 40, 34–44. [Google Scholar] [CrossRef]
- Vrieling, A.; Meroni, M.; Darvishzadeh, R.; Skidmore, A.K.; Wang, T.; Zurita-Milla, R.; Oosterbeek, K.; O’Connor, B.; Paganini, M. Vegetation phenology from Sentinel-2 and field cameras for a Dutch barrier island. Remote Sens. Environ. 2018, 215, 517–529. [Google Scholar] [CrossRef]








| Satellite Sensor | Revisit Interval (day) | Spatial Resolution (m) | Swath Width (km) | Radiometric Resolution (bit) | Blue (μm) | Green (μm) | Red (μm) | Near-Infrared (μm) |
|---|---|---|---|---|---|---|---|---|
| Gaofen-2 PMS | 4 | 4 | 45 | 10 | 0.45–0.52 | 0.52–0.59 | 0.63–0.69 | 0.77–0.89 |
| Sentinel-2 MSI | 5 | 10 | 290 | 12 | 0.46–0.52 | 0.54–0.58 | 0.65–0.68 | 0.79–0.90 |
| Gaofen-1 WFV | 4 | 16 | 800 | 10 | 0.45–0.52 | 0.52–0.59 | 0.63–0.69 | 0.77–0.89 |
| Landsat-8 OLI | 16 | 30 | 185 | 12 | 0.45–0.52 | 0.53–0.60 | 0.63–0.68 | 0.85–0.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Zhang, Z.; Yu, W.; Zhao, P.; Yue, J.; Deng, Y.; Geng, J. How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images. Remote Sens. 2021, 13, 2716. https://doi.org/10.3390/rs13142716
Xu K, Zhang Z, Yu W, Zhao P, Yue J, Deng Y, Geng J. How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images. Remote Sensing. 2021; 13(14):2716. https://doi.org/10.3390/rs13142716
Chicago/Turabian StyleXu, Kaijian, Zhaoying Zhang, Wanwan Yu, Ping Zhao, Jibo Yue, Yaping Deng, and Jun Geng. 2021. "How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images" Remote Sensing 13, no. 14: 2716. https://doi.org/10.3390/rs13142716
APA StyleXu, K., Zhang, Z., Yu, W., Zhao, P., Yue, J., Deng, Y., & Geng, J. (2021). How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images. Remote Sensing, 13(14), 2716. https://doi.org/10.3390/rs13142716