1. Introduction
The timing of planting, emergence, maturity, and harvest of crops affects the yield and long-term sustainability of croplands, thus tracking crop phenology has numerous interested parties from local to national levels. Remotely sensed data is an important data source for tracking crop phenology, and other attributes, from the field to global scales [
1]. For example, crop type, crop succession, and off-season management strategies can all be monitored using remote sensing data to inform large-scale management [
2]. Estimating crop phenology from remotely sensed data is difficult since crops do not follow the same patterns as natural vegetation. Harvest may happen when crops are still green, and multiple harvests in a single year will result in several “peaks” in greenness. Crop rotations result in different crop types year to year, which affects the relative greenness and derived phenology metrics [
3]. A primary limitation for improving satellite remote sensing-based crop phenology models is a lack of widespread ground truth data [
4]. Near-surface cameras used in the PhenoCam network offer a novel solution to this need for local-scale crop management information. Images from near-surface cameras can document the date of emergence, maturity, harvest, and tillage at the field scale with a daily temporal resolution [
5,
6].
Automatically extracting cropland attributes and phenological information from PhenoCam imagery is difficult. The primary method to estimate phenological metrics with PhenoCam data uses the direction and amplitude of a greenness metric (the green chromatic coordinate,
) of regions of interest in the camera field of view [
7,
8]. These metrics are well correlated with crop emergence and maturity, but cannot be used to directly identify other attributes such as crop type, flowering, or the presence of crop residue [
9]. Deep learning models provide a straightforward method for identifying information in images [
10], and can potentially identify phenological states directly as opposed to inferring them from relative greenness in the images. Studies have successfully used deep learning image classification models to identify and count animals [
11,
12], classify animal movement [
13], and identify the phenological stage [
14,
15], species [
16], or stressors [
17] of individual plants. Deep learning has been used previously with PhenoCams to identify images with snow cover with up to 98% accuracy [
18] and also with high accuracy to classify forest phenology [
19,
20].
In agriculture, deep learning classification of near-surface images, either from fixed or handheld cameras, has primarily been used for weed and crop disease detection [
21]. Few studies have used deep learning for the classification of crop and field attributes (e.g., [
22,
23,
24]) and to our knowledge no study has applied deep learning methods at cropland sites in the PhenoCam image archive [
25]. In addition to producing widespread ground truth data for large-scale models, automated classification of crop phenology would be beneficial in local-scale experiments of cropping practices. Recent studies have shown phenology from PhenoCams enables high-throughput field phenotyping and tracking a variety of crop characteristics [
6,
26].
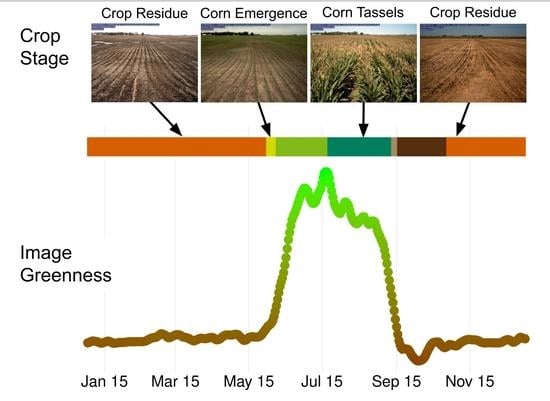
Here, we use images from 55 agricultural cameras in the PhenoCam network to build a classification model for identifying cropland phenological states. The database includes images with a variety of real-world conditions across several crop types (
Figure 1). We used a generalized classification scheme with 21 classes across 3 mutually exclusive categories, ranging from emergence to post-harvest residue. We also included classes for crop type and factors such as flooded or snow covered fields. Deep learning models designed for image classification do not have a temporal component, so we use a hidden Markov model in the classification post-processing to account for the temporal correlation of daily camera time series. Results show the feasibility of a daily, local-scale dataset of field states and phenological stages for agricultural research.
2. Methods
2.1. Data
We used PhenoCam images from agricultural sites to train an image classification model (
Figure S1, Table S1). To obtain a representative sample of images across all potential crops and crop stages, we used seasonal transition dates provided by the PhenoCam Network. Based on the transition date direction (either rising or falling) and threshold (10%, 25%, and 50%), we partitioned each calendar year into distinct periods of senesced, growth, peak, and senescing [
7]. We chose 50 random days from each site, year, and period, for a total of 8270 images. We annotated each image by hand using the imageant software into the 21 classes described below [
27].
Initial image classifications were organized into 21 classes across three categories of Dominant Cover, Crop Type, and Crop Status (
Table 1). The categories are each mutually exclusive such that any single image can be independently classified into a single class within each category. This allows finer grained classification given an array of Crop Types, and more flexibility in classifying crop phenological stages. For example, it would be informative for remote sensing models to know the exact date of crop emergence, but also that on the specified date and for several days to weeks after the feld is still predominantly bare soil. The first category, Dominant Cover, is the predominant class within the field of view. The Crop Type category represents the four predominant crops in the dataset (corn, wheat/barley, soybean, and alfalfa). Wheat and barley are combined into a single category as they are difficult to discern in images. The unknown Crop Type class is used during emergence when an exact identification is impossible. The other Crop Type class represents all other crops, including fallow fields, besides the four predominant ones. The stages of the Crop Status category are loosely based on BBCH descriptions [
28], but generalized to be applicable across a variety of crops and what is discernible. The Flowers stage is used solely for identifying reproductive structures for corn, wheat, or barely. No reproductive structures were discernible in images with Crop Types of Soybean, Alfalfa, or Other, thus images in those three Crop Type classes have no Crop Status annotations of Flowers.
After annotation we excluded some images based on low prevalence of some category combinations. For example, only 8 images had the combined combination of Soil, Unknown Plant, and Senescing for the Dominant Cover, Crop Type, and Crop status categories, respectively. When a unique combination of the three categories had less than 40 total images, all images representing that combination were excluded from the model fitting. This resulted in 255 annotated images, from the original 8270, being excluded. A total of 8015 annotated images were available for model fitting.
We used midday images in the annotation stage and leveraged more of the PhenoCam archive to increase sample size for model fitting. The 8015 images that we annotated represent the midday image for a single date, though phenocams record images up to every 30 min. For each annotated image date, we also downloaded all images between 0900 and 1500 local time, resulting in an additional 83,469 images. We applied the annotation of the midday image to all images of that date, resulting in 91,484 total images used in the model fitting. This allowed us to increase training image data by a factor of 10 with minimal effort, and include more variation in lighting conditions. While it is possible some of these non-midday images were annotated incorrectly (e.g., a blurry camera becoming cleared, or a field being plowed after midday), these are likely minimal and did not have a large effect on model accuracy [
12].
2.2. Image Classification Model
We used the VGG16 model in the Keras python package to classify images into the 21 classes (
Table 1) [
29,
30]. We evaluated several other models preconfigured in the Keras package and found VGG16 to be the best performing (see
Supplement). The model allows us to specify the hierarchical structure of the three categories, such that the predicted class probabilities for any image sum to one within each category. We held out 20% of the images as a validation set. The validation set included all images from three cameras: arsmorris2, mandani2, and cafboydnorthltar01 totaling 10,172 images. It also included 8124 randomly selected images from the remaining locations to obtain the full 20%. This resulted in a validation sample size of 18,296 and a training sample size of 73,188. The VGG16 classification model was trained fully, as opposed to using transfer learning [
12]. We experimented with transfer learning, where a pre-trained model is fine-tuned using our own data, but found that training the model fully had better results.
We resampled the 73,188 training images to 100,000 using weights proportional to the unique combinations among the three categories. For example, there were 4407 images annotated as Vegetation, Corn, and Flowers for the three categories, but only 1074 images annotated with Vegetation, Wheat, and Flowers. The images in the former class were given a lower weight in the resampling to reach 100,000 total training images. This allows for even sample sizes among classes and protects against the model being biased toward common classes. During model fitting the images are shuffled and transformed using random shifts and rotations such that the exact same image is never seen twice, which protects against over-fitting. We trained the model with an image resolution of 224 × 224 pixels using the Adams optimizer with a learning rate of 0.01 for 15 epochs, and an additional 5 epochs with a learning rate of 0.001.
2.3. Post-Processing
After fitting, the VGG16 model was used to classify approximately fifty-five thousand midday images from all agricultural PhenoCam sites, totalling approximately 170 site-years [
31]. These classified images were then put through a post-processing routine to produce a final classification for each day (
Table 2). First, for all dates marked as Snow in the Dominant Cover category, the Crop Type and Crop Status predictions were removed and gap-filled using linear interpolation from surrounding non-snow dates as long as the gap was 60 days or less. The reasoning behind this is during the constant snow cover of winter the crop, if any, likely remains unchanged. Next, any image marked as blurry was removed and the associated image date marked as missing across all three categories (Dominant Cover, Crop Type, Crop Status). Gaps of missing dates, up to 3 days, in the time series for each site were filled with a linear interpolation of the two bounding date probabilities for each of the remaining 18 classes. Probabilities across all dates were normalized across the remaining classes to account for removing the “Blurry” class. After this initial filtering the classification time series from the approximately fifty-five thousand images were input into an hidden Markov, and several additional post-processing steps, to produce the final time series.
Out-of-the box image classification models such as VGG16 have no temporal component. Every image is treated as an observation independent of temporally adjacent images. Thus, mis-classification of images can lead to noisy time series and improbable transitions between classes. To correct for this, we used a hidden Markov model (HMM) to reduce the day-to-day variation and remove improbable transitions [
32,
33,
34]. HMM’s are state-space models which combine a latent “true” state of a process with an observation model. The latent state evolves dynamically where every timestep is a discrete state which depends only on the state of the previous timestep, and where the probability of moving from one state to another is decided by a transition matrix. The observation model is a time series of the same length where every observed state depends only on the latent state of the same timestep. Given a latent state, all observed states have a non-zero probability of being observed.
We used two HMMs, one for Dominant Cover and another for Crop Status, each with a daily timestep. Only sequences with at least 60 continuous days, after the gap-filling from Snow and Blurry dates described above, were processed with the HMM. For the observation model within each HMM we used the direct output from the classification step, which for each image date consisted of probabilities of the image belonging to each class in the respective category. The transition matrix describing the probabilities of the hidden state changing states from one day to the next was created manually for each HMM (
Tables S2 and S3). The probability of the hidden state staying the same between two dates was set as the highest (0.90–0.95) in all cases. In this way, the hidden state only changes when there is strong evidence in the observation model, represented by continuous and high observed probabilities of a new state. Within the Dominant Cover category, transitions to other states besides the current one were set to equal values with one exception. Transition from Soil to Residue was set to 0 probability since residue can only be present after a crop has been harvested. Within Crop Status transitions between states were constrained to be biologically possible. Improbable transitions (e.g., from Emergence to Senescing in a single day) had probabilities of 0. Reproductive structures were not visible on all crops in images, so transitioning from the Growth stage directly to Senescing was allowed. Transitioning from either Senescing or Senesced to Growth was also allowed since this represents dormancy exit in overwintering crops such as winter wheat. Given observation probabilities and the transition matrices the most likely hidden state was predicted using the viterbi algorithm to produce the final time series across the Dominant Cover and Crop Status categories.
For the Crop Type category, the HMM methodology is less useful, since no day-to-day transitions between Crop Types are expected. Here, we used the HMM output of Crop Status to identify each unique crop sequence, defined as all dates between any two “No Crop” classifications. For each unique crop sequence, we identified the associated Crop Type with the highest total probability within that sequence, and marked the entire sequence as that Crop Type. In this step, we considered all Crop Type classes except “Unknown Plant”, which is used during the emergence stage when a plant is present but the exact type is unclear. This allows information later in the crop cycle, when Crop Type is more easily classified, to be propagated back to the emergence stage. Finally, we marked the final Crop Type as Unknown for some crop sequences in two instances: (1) when the length of a crop sequence is less than 60 days and the sequence was predominantly in the emergence stage, and (2) when the “No Cop” class was selected as the final Crop Type with the highest probability within a sequence. These two scenarios tended to occur when volunteer plants are growing sparsely on an otherwise bare field.
2.4. Evaluation
We calculated three metrics to evaluate the performance of the image classifier: precision, recall, and the
F1 score. All three metrics are based on predictions being classified into four categories of true positive (
), false positive (
), true negative (
), and false negative (
). Precision is the probability that an image is actually class
i, given that the model classified it as class
i. Recall is the probability that an image will be classified into class
i, given that the image is actually class
i. The
F1 score is the harmonic mean of precision and recall. All three metrics have the range 0–1, where 1 is a perfect classification.
where
,
,
are the number of true-positive, false-positive, and false-negative classifications, respectively. Using only the midday images we calculated all three metrics for the 21 original classes to evaluate the performance of the VGG16 model. For this step, the predicted class for each category on a single date was the one with the maximum probability. We re-calculated all three metrics again after all post-processing steps. The second round of metrics does not include two classes removed during post-processing: the Blurry class across all categories and the Unknown Plant class in the Crop Type category.
Software packages used throughout the analysis include Keras v2.4.3 [
30], TensorFlow v2.2.0 [
35], Pandas v1.0.4 [
36], NumPy v.1.20.2 [
37], and Pomegranate v0.14.5 [
38] in the Python programming language v3.7 [
39]. In the R language v4.1 [
40], we used the zoo v1.8.0 [
41], tidyverse v1.3.1 [
42], and ggplot2 v3.3.5 [
43] packages. All code for the analysis, as well as the final model predictions, are available in a Zenodo repository [
44].
3. Results
The overall
F1 score, a summary statistic which incorporates recall and precision, was 0.90–0.92 for the training data across the three categories of Dominant Cover, Crop Type, and Crop Status (
Figure 2). The overall
F1 score for validation data, which was not used in the model fitting, was 0.83–0.85 for the three categories. In the Dominant Cover category, the vegetation class was the best performing overall with recall and precision of 0.97 and 0.93, respectively. Thus, the classification model has a strong ability to discern when the camera field of view is or is not predominantly vegetation. When vegetation is not dominant, the classifier is still moderately accurate, though there is confusion between soil and residue classes indicated by their recall scores (0.64–0.68). The precision of soil and residue was 0.61 and 0.82 for validation data, indicating that the classifier leaned toward residue.
Excluding the blurry class, the worst precision for Crop Type was soybeans, with a precision of 0.53 on the validation data. This indicates that if an image was classified as Soybean, then there is a 53% chance it is actually soybean. The recall for Soybean was high, 0.97 with the validation data, indicating that there is a high amount of false positives from non-Soybean images being classified as Soybean. Conversely, the Wheat/Barley and Other classes have high precision (0.86 and 0.86, respectively), and low recall (0.60 and 0.65, respectively). This indicates a high amount of false negatives, where images of Wheat/Barley and Other Crop Types are being classified as other Crop Type classes (
Figures S2 and S3).
The blurry class had low recall and precision across all three categories, with values of 0.29–0.43 and 0.33–0.50 for validation data recall and precision, respectively. Combined with training data recall scores of 1.0, this indicates likely over-fitting of the blurry class in the classification model. We did not attempt to improve this further since the blurry image prevalence was extremely low. Additionally, when images were marked as blurry in the final dataset, the final state was interpolated in the HMM post-processing step by accounting for the surrounding images.
Figure 3 shows the classification statistics after post-processing of the image time series, where the HMM was used for Dominant Cover and Crop Status and the Crop Type was set to the highest total probability within any single crop series. The blurry class is not shown here since it was removed in the post-processing routine. The Unknown Plant class for Crop Type is also excluded since in the post-processing, the Crop Type category is assigned to the highest probability class seen in each crop sequence, thus performance metrics for the Unknown Plant class would be uninformative.
The validation data performance metrics after the post-processing steps either improved or remained the same across all classes except four: the Snow class in Dominant Cover, the Alfalfa Crop Type, and Flowers and Senescing Crop status classes. Overall F1 scores using validation data increased from 0.85 to 0.88, 0.83 to 0.91, and 0.85 to 0.86 for Dominant Cover, Crop Type, and Crop Status categories, respectively.
Next, we present four examples of the full classification and post-processing results using a single calendar year from four sites. They show the initial output of the classification model, as well as the capability of the HMM in removing high variation in the original VGG16 model prediction. The original VGG16 predictions are obtained by choosing the class with the maximum probability (MaxP) for each category and day. We compare them with insight gained from the full image time series available on the PhenoCam data portal
https://phenocam.sr.unh.edu, accessed 1 September 2021. For example, at the arsmorris2 site in central Minnesota, in the months March through June of 2020, there is uncertainty in whether the Dominant Cover of the field is Residue or Soil (
Figure 4A, MaxP). The HMM model resolved it to the Residue class for the three month period. From mid-June thru October there is high certainty that that vegetation is present, reflected in both the initial classifications (MaxP) and resulting HMM. The HMM model resolved the Crop Type as Corn (
Figure 4B). During June to October, the Crop Status progresses naturally through the different stages, and uncertainty arises only in October when fully senesced vegetation is confused with plant residue (
Figure 4C).
Cropping systems with multiple harvests per season are challenging for remote sensing models. There were multiple harvests for the bouldinalfalfa site in northern California for the year 2018. Here, an alfalfa field was persistent for the entire year with several harvests (
Figure 5A). During the intervals of regrowth after each harvest the Dominant Cover of the field was classified as Residue with emergence of an Unknown crop type (
Figure 5B). Once the plants matured then it was identified consistently as Alfalfa, which in the post-processing was back propagated in time for each crop sequence.
At the site cafcookeastltar01 in eastern Washington in the year 2018 there was a short residual crop of wheat in April (
Figure 6A). Since the plants were not allowed to grow into the summer, due to a new crop being planted, they were not positively identified and instead marked as Unknown Plant. From manual image interpretation, we know a crop of chickpeas was planted in May which grew until harvest in early September. Throughout the summer the model initially classifies this crop as wheat, soybean, or alfalfa. The post-processing correctly chose the Other Crop Type as the final class. In October and November there is confusion in the Dominant Cover category between soil and residue, even after post-processing. From the images, we can conclude there was likely no activity in the field during this time, thus confusion likely stems from a moderate amount of residue on the field combined with low light conditions of this northern (47.7° latitude) site.
Crops going into dormancy in the winter and resuming growth in the spring are accounted for in the post-processing routines as demonstrated by the Konza Agricultural site in the NEON network in 2017 (NEON.D06.KONA.DP1.00042,
Figure 7). A winter wheat crop (
Figure 7B), which was planted in the fall of 2016, resumed growth in February. The remainder of that crop life cycle proceeded normally until harvest in July (
Figure 7B). The crop type here is correctly classified as Unknown Plant by the classifier from January thru March here, since the plants were relatively small at this time. The correct classification of wheat began in March when the plants were large enough to confidently identify, and this was propagated back to the initial emergence in 2016. Additionally, the primary field at this site was harvested at the end of June 2017 (as seen in the original images), though the classification model indicated it happened mid-July (
Figure 7A). This was due to the foreground plants being removed in mid-July, while the primary field was harvested in June.
4. Discussion
Daily images from the PhenoCam network contain a wealth of information beyond just vegetation greenness, and here we showed they are also a novel source of cropland phenological information. Using a deep learning-based image classification model, we identified the daily field state, crop type and phenological state from PhenoCam images in agricultural fields. Since mainstream classification models do not have a temporal component we applied a hidden Markov model as a post-classification smoothing method which accounts for temporal correlation. This improved classification metrics and removed improbable transitions. Improvements would be beneficial to better classify field and crop states outside the primary growing season, and to better account for crops which go through a period of dormancy.
The classification model here was developed to simultaneously identify several crop and field attributes and has a variety of potential uses. For example, the United States Department of Agriculture currently monitors crop status throughout the USA using surveys [
45]. An array of PhenoCams positioned in representative fields could enable a real-time crop status monitoring system using the methodologies outlined here. Remote sensing models for monitoring crop progression would benefit from the large temporal and spatial extent of PhenoCams in agricultural fields as a source of verification data [
4]. The on-the-ground daily crop status data could also be used to parameterize or validate earth system models, where crop phenology is a primary source of error for crop yields [
46].
Numerous studies have used
from PhenoCams to study various biological processes [
25,
47]. Yet compressing images down to a single greenness index discards large amounts of information, especially in agricultural fields which are constantly managed [
9]. Our approach here allows us to extract more relevant data from images, such as the crop type or the state of the field after vegetation is removed. These image-based metrics of crop type and stage from PhenoCam time series can complement
as opposed to replacing it though. For example,
, and other greenness metrics, can be used to derive the date of peak greenness or the rate of greenup or greendown, which reflects important plant properties not present in our image classification approach such as water stress and plant vigor [
6,
26,
48]. Added insight from complementary metrics enrich interpretation and offer decision-makers flexibility in crop management [
9].
Most studies using deep learning methods to identify cropland attributes use satellite or aerial imagery [
21], though several studies have used near-surface imagery similar to the work here. Yalcin [
22] used a CNN to classify crop types and had
F1 scores of 0.74–0.87. Han et al. [
23] used a CNN to classify development stages in rice with
F1 scores ranging from 0.25–1.0. The high accuracy seen here and in other studies shows the capability of tracking crop and field attributes with near-surface cameras. This approach is advantageous since the cameras are not affected by cloud cover and, after initial installation, do not have significant labor costs.
We identified several areas of our approach which could be improved. Firstly, the VGG16 model used here could be replaced with either a more advanced or a customized neural network model. Though the initial accuracy of the VGG16 model was relatively high, it was originally designed for classification of common objects as opposed to croplands. It could likely be improved through model customization or fine-tuning of parameter estimation. Improving the initial image classification would improve the final results without any other adjustments to the post-processing routines.
Our approach here worked best during continuous periods where crops were present on the field. Once crops were removed, the dominant cover state could be difficult to discern due to soil, plant residue, and fully senesced plants having similar visual characteristics (
Figures S4–S11). Improvements could potentially be made here by using a zoomed-in or cropped photo of the field. Since the images were compressed from their full resolution to 224 × 224 pixels, it is likely important details were lost. Han et al. [
23] showed that zoomed-in images, used simultaneously with full resolution images in a custom neural network model, greatly improved accuracy of rice phenology classification. Using zoomed-in images may also help with identifying the reproductive structures of crops. Though this may be limited by camera placement since even during manual annotation we could only identify reproductive structures of corn, wheat, and barley. Residue versus Soil classification may also be improved by classifying the amount of residue (e.g., the fractional cover of plant residue or soil) as opposed to using two distinct classes.
Our use of an HMM is an ideal solution to account for temporal correlation in the classified image time series. The progression of crops at a daily time step is constrained by plant biology, and these constraints are easily built into the HMM using the transition matrix. Additionally, the predicted probabilities from the classification model can be used directly in the HMM observation model, resulting in a straightforward data workflow. Since we used a basic HMM we had to create separate models for the Dominant Cover and Crop Status categories, which resulted in occasional inconsistencies. For example, the Dominant Cover HMM may occasionally identify a time period as being predominantly vegetation, while the Crop Status HMM identifies the same time period as having no crop present (
Figure 7). A multi-level, or layered, HMM may be able to overcome this by modelling the joint probabilities of classes across the two categories [
49]. Temporal segmentation, a newer deep learning approach which is under active development, could model the joint probability of classes across the different categories in addition to having better performance than seen here [
50]. A downside to temporal segmentation is that it would require fully annotated training sequences (i.e., annotations for all images in a year for numerous sites) as opposed to the random selection of training images used here.
We observed some mis-classifications when field management activities are not uniformly applied to all parts of a field in the camera field of view. In the NEON-KONA example (
Figure 7), the final classification showed vegetation present due to foreground plants remaining even though the primary field was harvested. This could be improved by having a pre-processing step which identifies distinct agricultural fields within the camera field of view. Each agricultural field could then be classified independently. This would also allow the inclusion of PhenoCam sites focused on one to several experimental plots, which were excluded from this study. This step could be done automatically through image segmentation models, or manually as in the region of interest (ROI) identification in the current PhenoCam Network data workflow [
7].
Instead of discarding blurry or obscured images, we accounted for them directly in the modelling. This is ideal since real-time applications must account for such images without human intervention. The Blurry class across all three categories had high performance metrics for the training images, but with validation images it had the lowest performance among all classes. There are two possibilities for this low performance of this class. One is that the classification model was confident in classifying some partially obscured images as non-blurry where the human annotator was not (
Figures S12–S13). Second was the low sample size of the blurry image class, which had less than 70 total midday images. This likely resulted in the over-fitting of the blurry class on the training images and resulting low performance among validation images. Obtaining more PhenoCam images which are blurry or where the field of view was obscured in some way would be beneficial, and could be obtained from the numerous non-agricultural sites. Regardless, the low accuracy of blurry images had little effect on the final results, since the final classification of any single day is determined by the joint classifications of all surrounding days in the post-processing.