1. Introduction
Hyperspectral image is a three-dimensional image, which is captured by some aerospace vehicles carrying hyperspectral imagers. Hyperspectral images contain rich spectral–spatial information. Each sample of hyperspectral images has hundreds of spectral bands, and each band has hundreds of reflection information, which enables hyperspectral images to play a great role in military target detection, agricultural production, water quality detection, mineral exploration and other aspects [
1,
2,
3,
4]. Researchers have carried out a significant amount of useful research using the unique characteristics of hyperspectral images, for example, using the spectral information of hyperspectral images to detect the information of the earth’s surface [
5,
6]. Hyperspectral image classification is based on different kinds of substances with different spectral curves. Each category corresponds to some specific samples, and each sample also has its own unique spatial–spectral characteristics. However, there are two common problems in hyperspectral image classification: (1) using small sample training in hyperspectral image classification will affect the performance of the model and reduce the generalization ability of the model and (2) in the case of small samples, if we can fully extract spatial spectral features and improve the classification performance.
In the early stage of studying hyperspectral image classification, tools such as support vector machine [
7] and polynomial logistic regression [
8] are mainly used. The definition of support vector machine is the linear classifier with the largest interval in the feature spatial. Its learning strategy is to maximize the interval. Because hyperspectral images also contain a large number of nonlinear features, SVM cannot extract the non-linear features in hyperspectral images well. Although spectral information can be used for classification, the classification performance will be better if spatial information is fully used on the basis of spectral information. In order to further improve the classification performance, super-pixel sparse representation and multi-core learning are also proposed [
9,
10,
11]. A multi-core model has more flexibility and stronger feature mapping ability than single kernel function, but the algorithm of multi-core learning is more complex, inefficient and needs more memory and time.
Using deep learning technology can automatically extract the nonlinear and hierarchical features of hyperspectral images. For example, image classification [
12], semantic segmentation [
13] and target detection [
14] in computer vision tasks, information extraction [
15], machine translation [
16], question answering system [
17] in natural language processing and image classification. have made great progress with the help of deep learning technology. As a typical classification task, hyperspectral image classification, as a result of the progress of deep learning technology, the classification accuracy has also been greatly improved. So far, a great deal of exploration on extracting spectral–spatial features of hyperspectral images has been carried out. Some typical feature extraction methods [
18], such as the structural filtering method [
19,
20,
21] and morphological contour method [
22,
23,
24], random field method [
25,
26], sparse representation method [
27,
28] and segmentation method [
29,
30,
31] have been proposed. Compared with the traditional feature extraction method based on manual production, the deep learning method is an end-to-end method, which can learn useful features automatically from a large number of hyperspectral data through a multi-layer network. At present, the methods for extracting hyperspectral image features using depth learning include stack automatic encoder (SAE) [
32], depth belief network (DBN) [
33], CNNs [
34], recursive neural network [
35,
36] and graph convolution network [
37].
In [
33], Chen introduced the stacked automatic encoder (SAE) to extract important features. Tao [
38] extracted spectral–spatial features using two sparse SAE. Depth automatic coding will reduce the dimension of the input data. Its dimension reduction is different from PCA. Depth automatic coding is more complex because it carries out nonlinear operation. A new depth automatic encoder (DAE) was proposed by [
39] who designed a new collaborative representation method to process small training sets, which can obtain more useful features from the neighborhood of target pixels in hyperspectral images. Zhang [
40] used a recursive automatic encoder (RAE) and weighted method to fuse the extracted spatial information. In [
34], a depth belief network (DBN) was used for hyperspectral image classification. Although these methods can be classified, they are one-dimensional classification methods with poor classification performance. Hu [
41] proposed to directly extract the spectral features of hyperspectral images by using a one-dimensional CNN model and classified them by using the extracted spectral features; the model has five layers. Li [
42] proposed a new method for classifying hyperspectral images using pixels.
For hyperspectral image classification tasks, the 2D-CNNs model can directly extract spatial information. Some features in hyperspectral images are highly similar. In [
43], a depth two-dimensional CNNs model based on depth hash neural network (DHNN) is proposed. The proposed model can effectively learn the features with high similarity in hyperspectral images. Because hyperspectral images are high-dimensional data, it is necessary to reduce the dimension before classification, and then learn the spatial information in hyperspectral image samples through 2D-CNNs. Chen [
44] et al. proposed a method of acquiring spatial–spectral information of hyperspectral images and feature fusion using 2D-CNNs, which is based on deep neural network (DNN). The different region convolution neural network (DRCNN) to classify hyperspectral images was proposed by [
45]. The input of different regions is used to learn the context features of different regions, and better classification results are obtained. Zhu et al. [
46] proposed to introduce deformable convolution into the network to classify hyperspectral images, and adaptively adjust the receptive field size of the activation unit of a convolution neural network to effectively reflect the complex structure of hyperspectral images.
Hyperspectral image classification using 3D-CNNs has better performance, because both 1D-CNNs and 2D-CNNs cannot extract spatial feature information and spectral feature information at the same time. When using small training samples for hyperspectral image classification, the 3D-CNNs model is a more effective classification method, because it can capture the spatial–spectral information of hyperspectral images at the same time. Ding et al. [
47] proposed a convolutional neural network based on diverse branch modules (DBB). It enriches the spatial feature by combining branches with different scales and different complexities, including convolution sequence, multiscale convolution and average pooling. Thus, the feature extraction ability of single convolution is improved. Each branch uses convolution kernels with different scales to extract the spectral information of hyperspectral images, so as to improve the classification performance. Usually, convolution neural networks use pooling operations to reduce the size of feature map. This process is crucial to realize local spatial invariance and increase the receptive field of subsequent convolution. Therefore, the pooling operation should minimize the loss of information in the feature map. At the same time, computing and memory overhead should be limited. In order to meet these needs, Alexandros [
48] and others proposed a fast and efficient pooling method, 3D-Softpool, which can accumulate activation in an exponential weighted manner. Compared with other pooling methods, 3D-Softpool retains more information in the down sampling activation mapping. In hyperspectral image classification, finer down sampling can obtain more spatial feature information and can improve the classification accuracy.
A new network of dual branch dual attention mechanism (DBDA) is proposed in [
49]. One branch is used to extract spatial features and the other branch is used to extract spectral features. A spatial attention module is applied to spatial branches and a channel attention module is used to spectral branches. Some important spectral–spatial features can be captured by using the attention module, which helps to improve classification performance. The hyperspectral image classification methods based on 3D-CNNs can also be divided into two categories: (1) 3D-CNNs are utilized as a whole to extract the spectral–spatial features of hyperspectral images. In [
50], the depth feature extraction network of 3D-CNNs is proposed, which can capture spatial–spectral features at the same time. Some 3D-CNNs frameworks directly obtain the features of hyperspectral cubes without preprocessing and post-processing the input data. (2) Spectral features and spatial features are extracted and classified after feature fusion. In order to fully extract the spatial–spectral features of hyperspectral images from shallow layer to deep layer, a three-layer CNN is constructed in [
51]. Then, by fusing multi-layer spatial features and spectral features, more complementary information can be provided. Finally, the fused features and classifiers form a network, and the end-to-end performance optimization is carried out. Li et al. [
52] proposes deep CNN with double branch structure to extract spatial and spectral features.
The deep pyramid residual network (pResNet) proposed in [
53] can make full use of a large amount of information in hyperspectral images for classification, because the network can increase the dimension of feature mapping between layers. In [
54], an end-to-end fast dense spectral spatial convolution (FDSSC) hyperspectral image classification structure is proposed. Different convolution kernel sizes are used to extract spectral and spatial features respectively, and an effective convolution method is used to reduce the high dimension. Improving the running speed of the network can also effectively prevent over fitting. In order to avoid the loss of context information caused by using only one or several fixed windows as the input of hyperspectral image classification, an attention multi branch CNN structure using adaptive region search (RS–AMCNN) is proposed in [
55]. In [
56], a method for classifying hyperspectral images using multiscale super pixels and guided filter (MSS–GF) is proposed. MSS is used to obtain spatial local information from different scales in different regions, and a sparse representation classifier is used to generate classification maps of different scales in each region. This method can effectively improve the classification ability of hyperspectral images. Because RS–AMCNN can adaptively search the position of spatial window in local areas according to the specific distribution of samples, it can effectively extract edge information and evenly extract important features in each area. In [
57], a spectrum and context information set classification method based on Markov random field (MRF) is proposed. In order to make full use of deep features, a cascade MRF model is proposed to extract deep information. This method has good classification performance. In [
58], the proposed dual branch multi attention network (DBMA) extracts spectral spatial features, and uses the attention mechanism on both branches, which has a good classification effect. Sun et al. proposed a new method, low rank component induced spatial spectral kernel method based on patch, called lrcissk, for HSI classification. Through the low rank matrix recovery (LRMR) technology, the low rank features of the spectrum in HSI are reconstructed to explore more accurate spatial information, which is used to identify the homogeneous neighborhood pixels (i.e., centroid pixels) of the target [
59]. In [
60], this paper proposed a spectral–spatial feature tokenization transformer (SSFTT) method, which has a Gaussian weighted feature marker for function transformation, capturing spectral spatial features and advanced semantic features for hyperspectral image classification. Hong et al. proposed a method called invariant attribute profile (IAP) to extract invariant features from the spatial and frequency domain of hyperspectral images and classify hyperspectral images [
24]. Aletti, G. et al. proposed a new semi-supervised method for multilabel segmentation of HSI, which combines appropriate linear discriminant analysis and can be used to compare the similarity indexes of different spectra [
61]. Christos G. Bampis et al. proposed a graph driven image segmentation method. By developing the diffusion process defined on any graph, this method has less computational burden through experiments [
62].
The content of hyperspectral images is usually complex; many different substances show similar texture features, which means the performance of many CNN models cannot be brought into full play. Due to the existence of noise and redundancy in hyperspectral image data, standard CNNs cannot capture all features. In addition, when additional layers are added, the deeper CNNs architecture will also affect the convergence of the network and produce lower classification accuracy. In order to alleviate these problems, Ding et al. [
47] proposed DBB, which combines multiple branches with different scales and complexity to extract richer spectral feature information, including convolution sequence, multiscale convolution and average pooling. When using 3D-CNNs to extract features, the more layers of the network, the more complex the network structure will be, resulting in more parameters and more computing and memory overhead. In order to reduce information loss, 3D-Softpool [
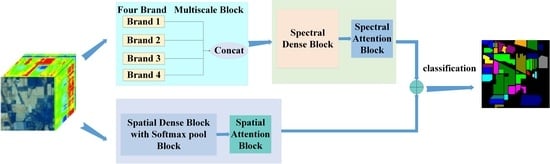
48] is used to extract spatial features, and 3D-Softpool can be cumulatively activated in an exponentially weighted manner. Compared with a series of other pooling methods, 3D-Softpool retains more information in down sampling activation mapping, which can effectively improve the performance and generalization ability of hyperspectral image classification. Inspired by DBB and 3D-Softpool methods, in order to fully extract spatial–spectral information and solve the problem of small sample over fitting, a spectral–spatial attention fusion method based on four branch multiscale blocks (FBMB) and sampling activation network based on 3D-Softpool module is proposed. The contributions of this study are as follows:
This paper proposes a FBMB structure different from other multi branches. The module enriches the feature spatial by combining multiple branches with different scales and complexity, and adds spectral attention blocks to each branch to further extract important spectral features. Finally, the extracted features of these branches are concatenated. The module can fully capture spatial–spectral features and improve the classification performance.
In the process of extracting spatial features, 3D-Softpool is introduced, and 3D-Softpool will be cumulatively activated in an exponential weighted manner. Compared with other pooling methods, 3D-Softpool retains more information in the down sampling activation mapping. A fusion method similar to dense connection is designed to extract the spectral and spatial features again to further improve the classification accuracy of hyperspectral images.
Experiments on four public data sets show that the experimental results of the proposed method for hyperspectral image classification are better than other advanced methods.
The rest of this paper is arranged as follows.
Section 2 introduces each part of the proposed method in detail.
Section 3 gives the experimental results and analysis.
Section 4 provides a discussion of the proposed method. In
Section 5, some conclusions are provided.
4. Discussion
Experiment 1: In order to verify the effectiveness of the proposed attention block, FBMB module, 3D-Softpool and dense connection, some ablation experiments were carried out on four hyperspectral images data sets. Only the modules to be tested in the network were deleted, and other parts remained unchanged.
Figure 11 shows the experimental results with or without specific modules. At the same time, it can be seen that the accuracy of the network reaches the highest after adding the spatial and spectral attention module. The reason is that the introduction of attention block to extract the features of hyperspectral images can adaptively allocate different weights, different spectral features and different spatial regions and selectively enhance the important features useful for classification, that is, increase the weight of some important features, which will help to improve the accuracy of classification. In addition,
Figure 11b shows the effectiveness of fusing spatial–spectral features of different scales. As shown in
Figure 11b, the FBMB module improves the classification accuracy of four data sets. This is because multiple branches can effectively extract spectral features of different scales, which is helpful to improve the classification accuracy. It is obvious from
Figure 11c that after removing the 3D-Softpool module, the OAs on the four data sets decrease significantly. 3D-Softpool block can reduce the loss of information in feature mapping and retain more information in down sampling activation mapping, so better classification results can be achieved on four data sets. As can be seen from
Figure 11d, if the dense connection is not used for the experiment, the OAS on the four data sets of IN, UP, SV and KSC decreases significantly. Using dense connection, all convolution layers can be connected, so that the spectral characteristic map output after convolution operation of each convolution layer is the input of all subsequent layers. This can ensure the maximum spectral information flow between network layers and make full use of spectral features. Therefore, better classification performance can be obtained on the four data sets of IN, UP, SV and KSC. As can be seen from
Figure 11d, if the dense connection is not adopted for the experiment, the OAs on the four data sets of IN, UP, SV and KSC decreases significantly. Using dense connection, all convolution layers can be connected, so that the spectral feature maps output by each convolution layer are the input of all subsequent layers. This can ensure the maximum spectral information flow between network layers and make full use of spectral features. Therefore, better classification performance can be obtained on the four data sets of IN, UP, SV and KSC.
Experiment 2:
Figure 12 is the schematic diagram of FBMB. In order to prove that our proposed FBMB can effectively improve the classification performance of hyperspectral image classification we carried out two groups of comparative experiments. Only the structure of FBMB was changed, and other structures remained unchanged. Some comparative experiments were carried out under the same conditions. The experimental results of the two groups of experiments were compared with that of the proposed method. In the first experiment, the convolution kernel size of all convolutions in FBMB were set to 7 × 7 × 7. We call the first group of comparative experiments the four-branch same scale block (FBSSB). In the second experiment, we used any three branches of the four branches B1, B2, B3 and B4 for experiments, which can have four combinations, namely C1 (B1, B2, B3), C2 (B1, B2, B4), C3 (B2, B3, B4) and C4 (B1, B3, B4). The experimental results of the three groups of comparative experiments on the four data sets of IN, UP, SV and KSC are shown in
Figure 13. As can be seen from
Figure 13, FBMB has the highest OA value and the best classification performance on the four data sets. The convolution kernel size of all convolutions in the first group of experiments is 7 × 7 × 7. The classification performance of this structure is not good. The size of convolution kernel is large, which leads to large number of parameters and slow operation in the experimental process. Compared with the combination of three branches, the feature extraction of four branches in FBMB is more sufficient and the classification performance is better.
Experiment 3: In order to further verify the performance of the proposed method, in comparison with other methods, the classification performance of different methods under different proportions of training samples was compared. In the experiment, the training ratios of IN, UP, KSC and SV were set to 1%, 5%, 10% and 15%, respectively. The experimental results are shown in
Figure 14. As can be seen from
Figure 14, when there are few training samples, the classification performance of CDCNN and SVM is relatively poor, and the classification performance of the proposed method is the best. With the increase in the number of samples, each method can obtain higher classification accuracy, but the classification accuracy of the proposed method can still be higher than that of other methods. This shows that this method has good generalization ability.
Experiment 4: Feature fusion merges the features extracted from the image into a complete image as the input of the next layer network, which can input more discriminative features for the next layer network. According to the order of fusion and prediction, feature fusion can be divided into early feature fusion and late feature fusion. Early feature fusion is a commonly used classical feature fusion method. For example, in an Inside–Outside Net (ION) [
65] or HyperNet [
66]), concatenation [
67] or addition operations are used to fuse certain layers. The feature fusion strategy in this experiment is an early fusion strategy that directly connects two spectral and spatial scale features. The two input features have the same size, and the output feature dimension is the sum of the two dimensions.
Table 10 shows the experimental results of whether to use the fusion strategy. It can be seen from
Table 10 that the OA values obtained on the four data sets after feature fusion are significantly higher than those obtained without feature fusion strategy. After feature fusion is used on each data set, the OA value obtained increases by more than 1.8%. The results show that the processing effect of feature fusion strategy on hyperspectral image classification is significantly improved compared with that without feature fusion strategy.