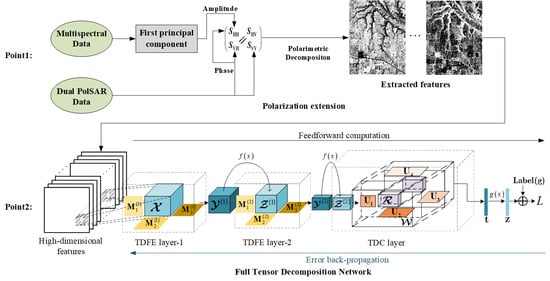
2.1. Multisource Data Fusion via Polarization Extension
Multispectral data and SAR data provide complementary features. The fusion of SAR data and multispectral data contributes to the better visual perception of objects and compensates for spatial information. Intensity hue saturation (IHS) and PCA methods are often used to merge multisensor data [
19]. Chavez investigated the feasibility of three methods for using panchromatic data to substitute spatial features of multispectral data (both statistically and visually) [
20]; Chandrakanth demonstrated the feasibility of the fusion of SAR data and multispectral data [
21]. A basic assumption concerning these methods is that the SAR amplitude is closely related to the intensity and principal component of multispectral images with high correlation coefficients. Therefore, SAR data can replace either of these two images while re-transforming data back into the original image space. Inspired by this assumption, we propose to consider an antithetical assumption that the principal component of multispectral images can be regarded as PolSAR data from the point of view of intensity, which yields the simulated SAR data in a certain polarization mode. Then the dual PolSAR data can be extended to construct synthetic quad PolSAR data.
For quad PolSAR data, each pixel is represented by a 2 × 2 complex matrix as follows
where
denotes the scattering factor of horizontal transmitting and vertical receiving polarization and the others have similar definitions. Here we demonstrate the fusion of multispectral data and dual PolSAR data generated in VH and VV polarization modes. Note that the reciprocity condition
is commonly assumed for quad PolSAR data, then we only need to construct the scattering factor
to form the quad PolSAR data. Firstly, we perform the registration of imageries collected by different sensors in advance. The overall fusion process is then illustrated in
Figure 1. On one hand, we extract the principal component of multispectral data using PCA. The extracted principal component represents the spatial information, which is used to generate the amplitude of
. On the other hand, the phase of
is simulated by using the phase of
instead. This is because they shared the same horizontal polarization transmitting antenna. Moreover, most of the distinguishable features (see
Table 1 below) for crop classification is irrelevant to the phase of scattering factor, and our experimental results show that the crop classification performance is insensitive to the phase of
. Finally, the simulated
is incorporated into the dual polarization data
and
to form synthetic quad PolSAR data, which enables us to extract higher dimensional features of the target through various polarimetric decomposition schemes. High-dimensional features extracted from synthetic quad PolSAR data provide more complementary information to multispectral data for accurate classification applications.
2.3. Full Tensor Decomposition Network
(1) Tucker decomposition-based feature extraction layer: When using the traditional neural network to classify high dimensional crop data, it is often necessary to perform dimensionality reduction operations to avoid the curse of dimensionality. Moreover, the extracted features from the data compressor may not be suitable for the classification model because the compressor and classification model are separately trained. We propose a Tucker decomposition-based feature extraction (TDFE) layer to extract hidden information from high-dimensional data. The TDFE layer can be used as the hidden layer of a tensor network, which performs the data compression or feature extraction for further processing.
The TDFE layer, different from the fully connected layer, uses tensor decomposition to realize the forward propagation. For a 3-way tensor
, a compressed feature tensor
can be obtained by transforming
as follows
where
are factor matrices,
performs the operation that multiplies each column fiber of
with
,
performs the operation that multiplies each row fiber of
with
, and
performs the operation that multiplies each tube fiber of
with
,
is the multi-linear transformed tensor and
is the activation function. Note that the outcome
is independent of the calculation order of mode product, each component
in tensor
is calculated by
where
,
,
are corresponding entries in matrices
,
and
. The forward propagation of the TDFE layer can be regarded as the inverse process of Tucker decomposition. The TDFE layer allows us to decompose the input tensor without destroying its coupling structure between each mode.
The factor matrices
are learned end-to-end by error backpropagation. When the TDFE layer is regarded as a hidden layer of a tensor neural network, we define the neuron error tensor
of the TDFE layer as
,where
L represents the loss function of the network. The update of each entry
is derived by finding the gradient
The updates of
and
are similar to that of
.
(2) Tucker decomposition-based classification layer: A flattened layer is always necessary for classical networks to vectorize the resulting features extracted by the hidden layer when classification or regression is performed. However, the structural information from the high-dimensional feature tensor will be discarded after the flattened layer. Instead, we propose to use a higher order weight tensor to project the feature tensor into a class vector without discarding the multimodal structure. The higher order weight tensor usually contains a large number of parameters, the update of which will lead to extremely high computational complexity. Here we proposed a Tucker decomposition-based classification (TDC) layer, which used the low-rank representation to replace the original weight tensor.
Figure 2 illustrates the structure of the TDC layer. Assuming that the 3-way feature tensor
is fed to the TDC layer, an output vector
can be generated by a 4-way weight tensor
, where
is the number of classes and each component
is calculated by the inner product of
and the slices of
where
denotes the inner product of two tensors, and
is the slice of
along the fourth way. In order to reduce the parameters of the TDC layer, the weight tensor
is decomposed and represented by a core tensor
and factor matrices
Then substituting (6) into (5) leads to
where
and
are 3-way smaller tensors of dimension
, since
can be always chosen far less than
. Note that the parameters of the TDC layer are converted to smaller core tensor
and the factor matrices
. Therefore, the number of parameters of the network can be greatly reduced.
When the TDC layer is used as the classification layer of a tensor neural network, the parameters of the TDC layer are learned end-to-end by error backpropagation. Let us define the neuron error vector
of the TDC layer as
, then the update of
and
are derived by finding the gradient
The update of each entry , , and is similar to that of .
(3) FTDN architecture and learning: Based on TDFE layer and TDC layer, we propose a full tensor decomposition network (FTDN) for high-dimensional data classification. The architecture of the FTDN is shown in
Figure 3, which contains two TDFE layers and one TDC layer. The input tensor sample
that corresponds to a certain pixel is formed by collecting all features of its local neighborhood pixels, where
is the number of features and
is the size of the neighborhood. For TDFE layer-1, the feedforward computation is straightforward, and its output feature tensor is denoted by
, where the superscript in the bracket denotes the layer index. For TDFE layer-2, the input tensor comes from the output of the previous layer, and the output feature tensor is denoted by
. For the TDC layer, the output vector
is transformed by a softmax layer to calculate the class posteriors
, where
represents the softmax function.
We now derive the error backpropagation-based learning of the FTDN. We see that a stochastic gradient descent learning for the FTDN is straightforward according to Equation (
3), (7) and (8), where the remained issue is to compute the neuron errors of each layer. As for layer TDC layer, the neuron error vector
depends on the specific loss function
L, which can be chosen from typical cross-entropy or mean square error between the network output and target label. Taking the cross entropy loss, for example, the neuron error vector
reads
, where
denotes the corresponding label vector. As for TDFE layer-2, the entries
of neuron error tensor
can be computed from
via the error backpropagation technique
Similarly, the error backpropagation from
to
is represented as