Synergistic Use of Citizen Science and Remote Sensing for Continental-Scale Measurements of Forest Tree Phenology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Assembling Citizen Science Observations of Phenology
- We sorted by the number of sites where each species was observed and selected all species that were observed at more than 30 sites.
- We sorted by the number of individuals observed at each site, and selected all observations made at sites with five or more species observed.
- Rule #1.
- We removed outlier observations by setting a priori thresholds to the date range in which acceptable observations could be made. This step was motivated by the observation that some NN phenophases that could be characterized as “spring phenophases” (e.g., flowers) were recorded as occurring in autumn, and “autumn phenophases” (e.g., colored leaves) were recorded as occurring in spring. The date range for acceptable observations in spring varied by species and phenophase, but was never earlier than DOY 50 or later than DOY 200. For autumn phenophases we accepted observations between DOY 200 and DOY 365.
- Rule #2.
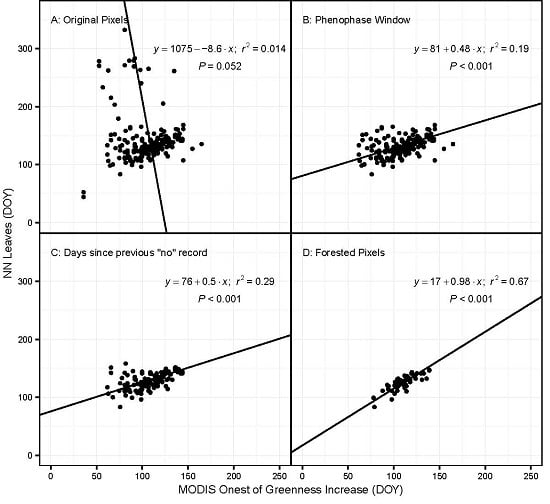
- All phenophase onset records were removed that were not preceded by a ‘no’ observation within the previous ~10 days. This effectively removed all observations made by volunteers who were not regularly visiting their plants and did not submit records to NN that were frequent enough to ensure a timely observation of an emerging phenophase. The specific threshold was identified by calculating the number of observations remaining after successive shortening of this window. The best threshold balances the need to use as many observations as possible with the need for regular observations of the same plant (Figure 1). The threshold number of days was 10 for most species; however, for some species this was lengthened after examining the effect on sample size (Table S1).
2.2. MODIS Phenology Product and Post-Processing
2.3. Statistical Analyses
3. Results
3.1. Model Performance for Poplar and Lilac
3.2. Species and Phenophase Model Variability
4. Discussion
4.1. Comparing Observations across Scales
4.2. Recommendations for Phenology-Focused Citizen Science Efforts
4.3. Could MODIS Be Used for High Throughput Phenotyping?
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DOY | Day of Year |
| MODIS | Moderate Resolution Imaging Spectrometer |
| NN | Natures Notebook |
| NPN | National Phenology Network |
| OGD | Onset of Greenness Decrease |
| OGI | Onset of Greenness Increase |
| OGMax | Onset of Greenness Maximum |
| OGMin | Onset of Greenness Minimum |
| LiDAR | Light Detection and Ranging |
| RADAR | RAdio Detection and Ranging |
References
- Richardson, A.D.; Keenan, T.F.; Migliavacca, M.; Ryu, Y.; Sonnentag, O.; Toomey, M. Climate change, phenology, and phenological control of vegetation feedbacks to the climate system. Agric. For. Meteorol. 2013, 169, 156–173. [Google Scholar] [CrossRef]
- Xu, L.; Myneni, R.B.; Chapin, F.S., III; Callaghan, T.V.; Pinzon, J.E.; Tucker, C.J.; Zhu, Z.; Bi, J.; Ciais, P.; Tommervik, H.; et al. Temperature and vegetation seasonality diminishment over northern lands. Nat. Clim. Chang. 2013, 3, 581–586. [Google Scholar] [CrossRef]
- Keenan, T.F.; Gray, J.; Friedl, M.A.; Toomey, M.; Bohrer, G.; Hollinger, D.Y.; Munger, J.W.; O’Keefe, J.; Schmid, H.P.; Wing, I.S.; et al. Net carbon uptake has increased through warming-induced changes in temperate forest phenology. Nat. Clim. Chang. 2014, 4, 598–604. [Google Scholar] [CrossRef]
- Myneni, R.B.; Keeling, C.D.; Tucker, C.J.; Asrar, G.; Nemani, R.R. Increased plant growth in the northern high latitudes from 1981 to 1991. Nature 1997, 386, 698–702. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Reiter, B.E. Changes in North American spring. Int. J. Climatol. 2000, 20, 929–932. [Google Scholar] [CrossRef]
- Cleland, E.E.; Chuine, I.; Menzel, A.; Mooney, H.A.; Schwartz, M.D. Shifting plant phenology in response to global change. Trends Ecol. Evolut. 2007, 22, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Chuine, I.; Belmonte, J.; Mignot, A. A modelling analysis of the genetic variation of phenology between tree populations. J. Ecol. 2000, 88, 561–570. [Google Scholar] [CrossRef]
- White, M.A.; de Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; van Leeuwen, W.J.D.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Liang, L.; Schwartz, M. Landscape phenology: An integrative approach to seasonal vegetation dynamics. Landsc. Ecol. 2009, 24, 465–472. [Google Scholar] [CrossRef]
- Liang, L.A.; Schwartz, M.D.; Fei, S.L. Validating satellite phenology through intensive ground observation and landscape scaling in a mixed seasonal forest. Remote Sens. Environ. 2011, 115, 143–157. [Google Scholar] [CrossRef]
- Melaas, E.K.; Friedl, M.A.; Zhu, Z. Detecting interannual variation in deciduous broadleaf forest phenology using Landsat TM/ETM plus data. Remote Sens. Environ. 2013, 132, 176–185. [Google Scholar] [CrossRef]
- Liang, L.; Schwartz, M.D.; Wang, Z.S.; Gao, F.; Schaaf, C.B.; Tan, B.; Morisette, J.T.; Zhang, X.Y. A cross comparison of spatiotemporally enhanced springtime phenological measurements from satellites and ground in a Northern U.S. Mixed forest. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7513–7526. [Google Scholar] [CrossRef]
- Keller, S.R.; Levsen, N.; Olson, M.S.; Tiffin, P. Local adaptation in the flowering-time gene network of balsam poplar, Populus balsamifera L. Mol. Biol. Evolut. 2012, 29, 3143–3152. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.S.; Levsen, N.; Soolanayakanahally, R.Y.; Guy, R.D.; Schroeder, W.R.; Keller, S.R.; Tiffin, P. The adaptive potential of Populus balsamifera L. to phenology requirements in a warmer global climate. Mol. Ecol. 2013, 22, 1214–1230. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, O.; Pyhajarvi, T.; Knurr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evolut. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- Hamunyela, E.; Verbesselt, J.; Roerink, G.; Herold, M. Trends in spring phenology of western European deciduous forests. Remote Sens. 2013, 5, 6159–6179. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Hanes, J.M. Intercomparing multiple measures of the onset of spring in eastern North America. Int. J. Climatol. 2009, 30, 1614–1626. [Google Scholar] [CrossRef]
- Craine, J.M.; Battersby, J.; Elmore, A.J.; Jones, A.W. Building EDENs: The rise of environmentally distributed ecological networks. Bioscience 2007, 57, 45–54. [Google Scholar] [CrossRef]
- Dickinson, J.L.; Zuckerberg, B.; Bonter, D.N. Citizen science as an ecological research tool: Challenges and benefits. Annu. Rev. Ecol. Evolut. Syst. 2010, 41, 149–172. [Google Scholar] [CrossRef]
- Cooper, C.B.; Hochachka, W.M.; Dhondt, A.A. The opportunities and challenges of citizen science as a tool for ecological research. In Citizen Science: Public Collaboration in Environmental Research; Cornell University Press: Ithaca, NY, USA, 2011; pp. 99–113. [Google Scholar]
- Denny, E.G.; Gerst, K.L.; Miller-Rushing, A.J.; Tierney, G.L.; Crimmins, T.M.; Enquist, C.A.F.; Guertin, P.; Rosemartin, A.H.; Schwartz, M.D.; Thomas, K.A.; et al. Standardized phenology monitoring methods to track plant and animal activity for science and resource management applications. Int. J. Biometeorol. 2014, 58, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Shirk, J.L.; Ballard, H.L.; Wilderman, C.C.; Phillips, T.; Wiggins, A.; Jordan, R.; McCallie, E.; Minarchek, M.; Lewenstein, B.V.; Krasny, M.E.; et al. Public participation in scientific research: A framework for deliberate design. Ecol. Soc. 2012, 17. [Google Scholar] [CrossRef]
- Bonney, R.; Shirk, J.L.; Phillips, T.B.; Wiggins, A.; Ballard, H.L.; Miller-Rushing, A.J.; Parrish, J.K. Next steps for citizen science. Science 2014, 343, 1436–1437. [Google Scholar] [CrossRef] [PubMed]
- Bird, T.J.; Bates, A.E.; Lefcheck, J.S.; Hill, N.A.; Thomson, R.J.; Edgar, G.J.; Stuart-Smith, R.D.; Wotherspoon, S.; Krkosek, M.; Stuart-Smith, J.F.; et al. Statistical solutions for error and bias in global citizen science datasets. Biol. Conserv. 2014, 173, 144–154. [Google Scholar] [CrossRef]
- Fitzpatrick, M.C.; Preisser, E.L.; Ellison, A.M.; Elkinton, J.S. Observer bias and the detection of low-density populations. Ecol. Appl. 2009, 19, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Delaney, D.G.; Sperling, C.D.; Adams, C.S.; Leung, B. Marine invasive species: Validation of citizen science and implications for national monitoring networks. Biol. Invasions 2008, 10, 117–128. [Google Scholar] [CrossRef]
- Edgar, G.J.; Barrett, N.S.; Morton, A.J. Biases associated with the use of underwater visual census techniques to quantify the density and size-structure of fish populations. J. Exp. Mar. Biol. Ecol. 2004, 308, 269–290. [Google Scholar] [CrossRef]
- Fuccillo, K.K.; Crimmins, T.M.; de Rivera, C.E.; Elder, T.S. Assessing accuracy in citizen science-based plant phenology monitoring. Int. J. Biometeorol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Edgar, G.J.; Stuart-Smith, R.D. Ecological effects of marine protected areas on rocky reef communities—A continental-scale analysis. Mar. Ecol. Prog. Ser. 2009, 388, 51–62. [Google Scholar] [CrossRef]
- Danielsen, F.; Jensen, P.M.; Burgess, N.D.; Altamirano, R.; Alviola, P.A.; Andrianandrasana, H.; Brashares, J.S.; Burton, A.C.; Coronado, I.; Corpuz, N.; et al. A multicountry assessment of tropical resource monitoring by local communities. Bioscience 2014, 64, 236–251. [Google Scholar] [CrossRef]
- See, L.; Comber, A.; Salk, C.; Fritz, S.; van der Velde, M.; Perger, C.; Schill, C.; McCallum, I.; Kraxner, F.; Obersteiner, M. Comparing the quality of crowdsourced data contributed by expert and non-experts. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Crimmins, T.M.; Crimmins, M.A.; Bertelsen, C.D. Flowering range changes across an elevation gradient in response to warming summer temperatures. Glob. Chang. Biol. 2009, 15, 1141–1152. [Google Scholar] [CrossRef]
- Crimmins, T.M.; Weltzin, J.F.; Rosemartin, A.H.; Surina, E.M.; Marsh, L.; Denny, E.G. Focused campaign increases activity among participants in nature’s notebook, a citizen science project. Nat. Sci. Educ. 2014, 43, 64–72. [Google Scholar]
- Ganguly, S.; Friedl, M.A.; Tan, B.; Zhang, X.; Verma, M. Land surface phenology from MODIS: Characterization of the collection 5 global land cover dynamics product. Remote Sens. Environ. 2010, 114, 1805–1816. [Google Scholar] [CrossRef]
- Elmore, A.J.; Guinn, S.M.; Minsley, B.J.; Richardson, A.D. Landscape controls on the timing of spring, autumn, and growing season length in mid-Atlantic forests. Glob. Chang. Biol. 2012, 18, 656–674. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F. Cross-scalar satellite phenology from ground, Landsat, and MODIS data. Remote Sens. Environ. 2007, 109, 261–273. [Google Scholar] [CrossRef]
- Fisher, J.I.; Richardson, A.D.; Mustard, J.F. Phenology model from surface meteorology does not capture satellite-based greenup estimations. Glob. Chang. Biol. 2007, 13, 707–721. [Google Scholar] [CrossRef]
- Yue, X.; Unger, N.; Keenan, T.F.; Zhang, X.; Vogel, C.S. Probing the past 30-year phenology trend of US deciduous forests. Biogeosciences 2015, 12, 4693–4709. [Google Scholar] [CrossRef]
- Rosemartin, A.H.; Denny, E.G.; Weltzin, J.F.; Lee Marsh, R.; Wilson, B.E.; Mehdipoor, H.; Zurita-Milla, R.; Schwartz, M.D. Lilac and honeysuckle phenology data 1956–2014. Sci. Data 2015, 2, 150038. [Google Scholar] [CrossRef] [PubMed]
- Madritch, M.D.; Greene, S.L.; Lindroth, R.L. Genetic mosaics of ecosystem functioning across aspen-dominated landscapes. Oecologia 2009, 160, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.E.; Ustin, S.L. Effects of microtopography and hydrology on phenology of an invasive herb. Ecography 2009, 32, 860–870. [Google Scholar] [CrossRef]
- Frolking, S.; Milliman, T.; McDonald, K.; Kimball, J.; Zhao, M.S.; Fahnestock, M. Evaluation of the seawinds scatterometer for regional monitoring of vegetation phenology. J. Geophys. Res.-Atmos. 2006, 111. [Google Scholar] [CrossRef]
- Jones, M.O.; Kimball, J.S.; Jones, L.A.; McDonald, K.C. Satellite passive microwave detection of North America start of season. Remote Sens. Environ. 2012, 123, 324–333. [Google Scholar] [CrossRef]








| Species | Common Name | Life Form |
|---|---|---|
| Acer negundo | boxelder | Lower canopy tree |
| Acer rubrum | red maple | Upper canopy tree |
| Acer saccharum | sugar maple | Upper canopy tree |
| Betula papyrifera | paper birch | Upper canopy tree |
| Cercis canadensis | eastern redbud | Lower canopy tree |
| Cornus florida | flowering dogwood | Lower canopy tree |
| Cornus sericea | redosier dogwood | Shrub |
| Fagus grandifolia | American beech | Upper canopy tree |
| Forsythia × intermedia | forsythia | Shrub |
| Fraxinus americana | white ash | Upper canopy tree |
| Hamamelis virginiana | American witchhazel | Lower canopy tree |
| Liquidambar styraciflua | sweetgum | Upper canopy tree |
| Liriodendron tulipifera | tuliptree | Upper canopy tree |
| Populus tremuloides | quaking aspen | Upper canopy tree |
| Prunus persica | peach | Lower canopy tree |
| Prunus serotina | black cherry | Upper canopy tree |
| Prunus virginiana | chokecherry | Shrub |
| Quercus alba | white oak | Upper canopy tree |
| Quercus rubra | northern red oak | Upper canopy tree |
| Rosa rugosa | rugosa rose | Shrub |
| Sambucus nigra | black elderberry | Shrub |
| Symphoricarpos albus | common snowberry | Shrub |
| Syringa vulgaris | common lilac | Shrub |
| Syringa × chinensis | Red Rothomagensis lilac | Shrub |
| Tilia americana | American basswood | Upper canopy tree |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elmore, A.J.; Stylinski, C.D.; Pradhan, K. Synergistic Use of Citizen Science and Remote Sensing for Continental-Scale Measurements of Forest Tree Phenology. Remote Sens. 2016, 8, 502. https://doi.org/10.3390/rs8060502
Elmore AJ, Stylinski CD, Pradhan K. Synergistic Use of Citizen Science and Remote Sensing for Continental-Scale Measurements of Forest Tree Phenology. Remote Sensing. 2016; 8(6):502. https://doi.org/10.3390/rs8060502
Chicago/Turabian StyleElmore, Andrew J., Cathlyn D. Stylinski, and Kavya Pradhan. 2016. "Synergistic Use of Citizen Science and Remote Sensing for Continental-Scale Measurements of Forest Tree Phenology" Remote Sensing 8, no. 6: 502. https://doi.org/10.3390/rs8060502
APA StyleElmore, A. J., Stylinski, C. D., & Pradhan, K. (2016). Synergistic Use of Citizen Science and Remote Sensing for Continental-Scale Measurements of Forest Tree Phenology. Remote Sensing, 8(6), 502. https://doi.org/10.3390/rs8060502