An Improved RANSAC for 3D Point Cloud Plane Segmentation Based on Normal Distribution Transformation Cells
Abstract
:1. Introduction
2. Related Works
2.1. NDT Methods
2.2. RANSAC
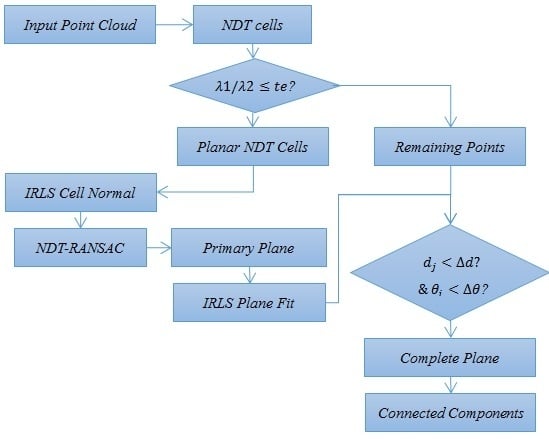
3. Proposed Methods
3.1. NDT Feature
- If , the distribution of the NDT cell is linear.
- If it is nonlinear and , the distribution of the NDT cell is planar.
- If no eigenvalue is times larger than another one, the distribution of the NDT cell is spherical.
| Algorithm 1. NDT Feature Calculation. |
| Input: P: an unorganized point cloud; : cell size; : a threshold to classify cells as planar or non-planar Initialize: ; // NDT cell list ; // planar NDT cell list ; // remaining points for each point in P Subdivide the space occupied by the scan into a grid of cubes ; Calculate the encoding of each point , which indicates the index of the cell; end for for each cell in Calculate the mean of the points in each cell; Calculate the covariance of each NDT cell; Eigen-decomposition for , get the eigenvalues and the corresponding eigenvectors of ; if Add cell into ; else if Add points into ; end if end for return |
3.2. NDT-RANSAC Plane Segmentation
| Algorithm 2. NDT-RANSAC Plane Segmentation. |
| Input: : planar NDT cell list : non-planar points Initialize: : plane surface while randomly sample a cell c; then, its gravity and normal are used as plane parameter // Calculate the support set for each cell in ; ; if add to ; end if If then ; recompute from Equation (3) using ; end if ; end while refine plane ; for each point in ; ; if and add points in plane ; End if end for |
3.3. Probabilities
3.4. IRLS Normal Calculation and Plane Fitting
| Algorithm 3. IRLS Plane Fitting. |
| Input: : points on a detected plane : maximum number of iterations : threshold to terminate iterations Initialize: ; // last normal Calculate the mean of the plane point cloud ; Calculate the covariance of the plane point cloud; Eigen-decomposition of , get the eigenvalues and the corresponding eigenvectors of ; n = ; // take as the normal parameter of the plane; for k = 1 to ; ; Calculate Welsch function ; ; ; Eigen-decomposition of , the corresponding eigenvector of the smallest eigenvalue is used as plane parameter ; ; if then Terminate iterations; end if end for |
3.5. Connected-Component Analysis
3.6. NDT Cell Size
4. Experimental Section
5. Discussion
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vosselman, G. Point cloud segmentation for urban scene classification. In Proceedings of the ISPRS International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Antalya, Turkey, 11–17 November 2013. [Google Scholar]
- Orthuber, E. 3D Building reconstruction from airborne LiDAR point clouds. In Proceedings of the ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Munich, Germany, 25–27 March 2015. [Google Scholar]
- Yang, B.; Dong, Z. A shape-based segmentation method for mobile laser scanning point clouds. ISPRS J. Photogramm. Remote Sens. 2013, 81, 19–30. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Li, D. A method based on an adaptive radius cylinder model for detecting pole-like objects in mobile laser scanning data. Remote Sens. Lett. 2016, 7, 249–258. [Google Scholar] [CrossRef]
- Li, L.; Li, D.; Zhu, H.; Li, Y. A dual growing method for the automatic extraction of individual trees from mobile laser scanning data. ISPRS J. Photogramm. Remote Sens. 2016, 120, 37–52. [Google Scholar] [CrossRef]
- Xiao, J.; Adler, B.; Zhang, H. 3D point cloud registration based on planar surfaces. In Proceedings of the IEEE Multisensor Fusion and Integration for Intelligent Systems, Hamburg, Germany, 13–15 September 2012. [Google Scholar]
- Poppinga, J.; Vaskevicius, N.; Birk, A.; Pathak, K. Fast plane detection and polygonalization in noisy 3D range images. In Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems, Nice, France, 22–26 September 2008. [Google Scholar]
- Rusu, R.B.; Marton, Z.C.; Blodow, N.; Dolha, M.; Beetz, M. Towards 3D point cloud based object maps for household environments. Robot. Auton. Syst. 2008, 56, 927–941. [Google Scholar] [CrossRef]
- Xiong, X.; Adan, A.; Akinci, B.; Huber, D. Automatic creation of semantically rich 3D building models from laser scanner data. Autom. Constr. 2013, 31, 325–337. [Google Scholar] [CrossRef]
- Previtali, M.; Scaioni, M.; Barazzetti, L.; Brumana, R. A flexible methodology for outdoor/indoor building reconstruction from occluded point clouds. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, II-3, 119–126. [Google Scholar] [CrossRef]
- Vaskevicius, N.; Birk, A.; Pathak, P.; Schwertfeger, S. Efficient Representation in Three-Dimensional Environment Modeling for Planetary Robotic Exploration. Adv. Robot. 2010, 24, 1169–1197. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, J.; Adler, B.; Zhang, H.; Zhang, J. Three-dimensional point cloud plane segmentation in both structured and unstructured environments. Robot. Auton. Syst. 2013, 61, 1641–1652. [Google Scholar] [CrossRef]
- Nguyen, A.; Le, B. 3D point cloud segmentation: A survey. In Proceedings of the IEEE Robotics, Automation and Mechatronics, Manila, Philippines, 12–15 November 2013. [Google Scholar]
- Vo, A.V.; Truong-Hong, L.; Laefer, D.F.; Bertolotto, M. Octree-based region growing for point cloud segmentation. ISPRS J. Photogramm. Remote Sens. 2015, 104, 88–100. [Google Scholar] [CrossRef]
- Borrmann, D.; Elseberg, J.; Kai, L.; Nüchter, A. The 3D Hough Transform for plane detection in point clouds: A review and a new accumulator design. 3D Res. 2011, 2, 1–13. [Google Scholar] [CrossRef]
- Rabbani, T.; van den Heuvel, F.A.; Vosselman, G. Segmentation of point clouds using smoothness constraint. In Proceedings of the International Archives of Photogrammetry, Remote Sensing & Spatial Information Sciences, Goa, India, 25–30 September 2006. [Google Scholar]
- Sampath, A.; Shan, J. Segmentation and Reconstruction of Polyhedral Building Roofs from Aerial LiDAR Point Clouds. IEEE Trans. Geosci. Remote Sens. 2010, 48, 1554–1567. [Google Scholar] [CrossRef]
- Tarsha-Kurdi, F.; Landes, T.; Grussenmeyer, P.; Koehl, M. Model-driven and data-driven approaches using LIDAR data: Analysis and comparison. In Proceedings of the Photogrammetric Image Analysis, Munich, Germany, 19–21 September 2007; pp. 87–92. [Google Scholar]
- Awwad, T.M.; Zhu, Q.; Du, Z.; Zhang, Y. An improved segmentation approach for planar surfaces from unstructured 3D point clouds. Photogramm. Rec. 2010, 25, 5–23. [Google Scholar] [CrossRef]
- Xu, B.; Jiang, W.; Shan, J.; Zhang, J.; Li, L. Investigation on the Weighted RANSAC Approaches for Building Roof Plane Segmentation from LiDAR Point Clouds. Remote Sens. 2015, 8, 5. [Google Scholar] [CrossRef]
- Torr, P.H.S.; Zisserman, A. MLESAC: A New Robust Estimator with Application to Estimating Image Geometry. Comput. Vis. Image Underst. 2000, 78, 138–156. [Google Scholar] [CrossRef]
- Wang, M.; Tseng, Y.H. Automatic Segmentation of Lidar Data into Coplanar Point Clusters Using an Octree-Based Split-and-Merge Algorithm. Photogramm. Eng. Remote Sens. 2010, 4, 407–420. [Google Scholar] [CrossRef]
- Su, Y.-T.; Bethel, J.; Hu, S. Octree-based segmentation for terrestrial LiDAR point cloud data in industrial applications. ISPRS J. Photogramm. Remote Sens. 2016, 113, 59–74. [Google Scholar] [CrossRef]
- Aijazi, A.K.; Checchin, P.; Trassoudaine, L. Segmentation Based Classification of 3D Urban Point Clouds: A Super-Voxel Based Approach with Evaluation. Remote Sens. 2013, 5, 1624–1650. [Google Scholar] [CrossRef]
- Biber, P.; Strasser, W. The normal distributions transform: A new approach to laser scan matching. In Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems, Las Vegas, NV, USA, 27–31 October 2003. [Google Scholar]
- Magnusson, M.; Lilienthal, A.; Duckett, T. Scan registration for autonomous mining vehicles using 3D-NDT: Research Articles. J. Field Robot. 2007, 24, 803–827. [Google Scholar] [CrossRef]
- Green, W.R.; Grobler, H. Normal distribution transform graph-based point cloud segmentation. In Proceedings of the Pattern Recognition Association of South Africa and Robotics and Mechatronics International Conference, Port Elizabeth, South Africa, 26–27 November 2015. [Google Scholar]
- Stoyanov, T.; Magnusson, M.; Almqvist, H.; Lilienthal, A.J. On the accuracy of the 3D Normal Distributions Transform as a tool for spatial representation. In Proceedings of the IEEE International Conference on Robotics & Automation, Shanghai, China, 9–13 May 2011. [Google Scholar]
- Magnusson, M. The Three-Dimensional Normal-Distributions Transform—An Efficient Representation for Registration, Surface Analysis, and Loop Detection. Renew. Energy 2012, 28, 655–663. [Google Scholar]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1980, 24, 381–395. [Google Scholar] [CrossRef]
- Schnabel, R.; Wahl, R.; Klein, R. Efficient RANSAC for Point-Cloud Shape Detection. Comput. Graph. Forum 2007, 26, 214–226. [Google Scholar] [CrossRef]
- Raguram, R.; Chum, O.; Pollefeys, M.; Matas, J.; Frahm, J.M. USAC: A universal framework for random sample consensus. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2022–2038. [Google Scholar] [CrossRef] [PubMed]
- Poreba, M.; Goulette, F. RANSAC algorithm and elements of graph theory for automatic plane detection in 3D point clouds. Arch. Photogramm. 2012, 24, 301–310. [Google Scholar]
- Fujiwara, T.; Kamegawa, T.; Gofuku, A. Plane detection to improve 3D scanning speed using RANSAC algorithm. In Proceedings of the Industrial Electronics and Applications, Melbourne, Australia, 19–21 June 2013; pp. 1863–1869. [Google Scholar]
- Weinmann, M.; Jutzi, B.; Mallet, C. Semantic 3D scene interpretation: A framework combining optimal neighborhood size selection with relevant features. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, II-3, 181–188. [Google Scholar] [CrossRef]
- Marden, S.; Guivant, J. Improving the performance of ICP for real-time applications using an approximate nearest neighbour search. In Proceedings of the Australasian Conference on Robotics and Automation, Wellington, New Zealand, 3–5 December 2012. [Google Scholar]
- Fayez, T.K.; Landes, T.; Grussenmeyer, P. Extended RANSAC algorithm for automatic detection of building roof planes from LiDAR data. Photogramm. J. Finl. 2008, 21, 97–109. [Google Scholar]
- Aftab, K.; Hartley, R. Convergence of Iteratively Re-weighted Least Squares to Robust M-Estimators. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 5–9 January 2015. [Google Scholar]
- Zhang, Z. Parameter estimation techniques: A tutorial with application to conic fitting. Image Vis. Comput. 1997, 15, 59–76. [Google Scholar] [CrossRef]
- Rooms UZH Irchel Dataset. Available online: http://www.ifi.uzh.ch/en/vmml/research/datasets.html (accessed on 12 October 2016).
- Pomerleau, F.; Liu, M.; Colas, F.; Siegwart, R. Challenging data sets for point cloud registration algorithms. Int. J. Robot. Res. 2012, 31, 1705–1711. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Cheng, L.; Li, M.; Wang, Y.; Xia, N. Three-Dimensional Reconstruction of Building Roofs from Airborne LiDAR Data Based on a Layer Connection and Smoothness Strategy. Remote Sens. 2016, 8, 415. [Google Scholar] [CrossRef]










| Test Sites | Length (m) | Width (m) | Height (m) | Points | Average Density (Points/m2) |
|---|---|---|---|---|---|
| Room-1 | 7.0 | 5.4 | 3.2 | 11,050,391 | 91,352 |
| Room-2 | 8.6 | 10.2 | 2.8 | 367,602 | 1486 |
| Room-3 | 7.7 | 9.7 | 2.8 | 367,602 | 1745 |
| Dataset | Method | Correctness | SR | Completeness | Quality | Time (s) |
|---|---|---|---|---|---|---|
| Room-1 | RANSAC | 37.3% | 62.7% | 52.5% | 26.3% | 333.72 |
| NDT-RANSAC | 98.3% | 0 | 85.0% | 83.8% | 7.43 | |
| Room-2 | RANSAC | 45.9% | 54.1% | 70.1% | 38.3% | 5.57 |
| NDT-RANSAC | 90.4% | 0 | 98.1% | 88.7% | 5.03 | |
| Room-3 | RANSAC | 49.5% | 50.5% | 71.4% | 40.9% | 5.29 |
| NDT-RANSAC | 88.5% | 0 | 94.6% | 84.3% | 2.46 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Yang, F.; Zhu, H.; Li, D.; Li, Y.; Tang, L. An Improved RANSAC for 3D Point Cloud Plane Segmentation Based on Normal Distribution Transformation Cells. Remote Sens. 2017, 9, 433. https://doi.org/10.3390/rs9050433
Li L, Yang F, Zhu H, Li D, Li Y, Tang L. An Improved RANSAC for 3D Point Cloud Plane Segmentation Based on Normal Distribution Transformation Cells. Remote Sensing. 2017; 9(5):433. https://doi.org/10.3390/rs9050433
Chicago/Turabian StyleLi, Lin, Fan Yang, Haihong Zhu, Dalin Li, You Li, and Lei Tang. 2017. "An Improved RANSAC for 3D Point Cloud Plane Segmentation Based on Normal Distribution Transformation Cells" Remote Sensing 9, no. 5: 433. https://doi.org/10.3390/rs9050433
APA StyleLi, L., Yang, F., Zhu, H., Li, D., Li, Y., & Tang, L. (2017). An Improved RANSAC for 3D Point Cloud Plane Segmentation Based on Normal Distribution Transformation Cells. Remote Sensing, 9(5), 433. https://doi.org/10.3390/rs9050433