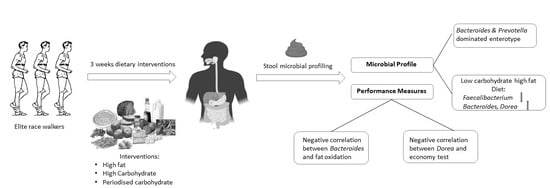
The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers
Abstract
:- ➢
- The gut microbiota profiles of endurance race walkers could be separated into two principal “enterotypes.”
- ➢
- The LCHF diet is associated with a significant reduction in the relative abundance of Faecalibacterium spp. and an increase in the relative abundance of Bacteroides and Dorea spp.
- ➢
- The relative abundance of some bacterial taxa was correlated with measures of athlete performance and metabolic capacity.
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. Stool Microbiota Analysis
2.3. Ethics Approval, Clinical Trial Registration and Availability of Data
3. Results
3.1. Bacteroides- or Prevotella-Enterotypes Are Predominant in Elite Race Walkers
3.2. HCHO and PCHO Diets Result in Subtle but Distinct Alterations in Firmicutes Lineages
3.3. The Low Carbohydrate High Fat Diet Results in More Profound Effects on the Gut Microbiota
3.4. Bacteroides and Dorea spp. Abundances Are Negatively Correlated with Athlete Performance Measures Following Consumption of the LCHF Diet
3.5. Is There a LCHF × Enterotype Interaction?
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E.; et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 15, R66. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, S.F.; Murphy, E.F.; O’sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 2017, 5, 98. [Google Scholar] [CrossRef] [Green Version]
- Burke, L.M.; Hawley, J.A.; Jeukendrup, A.; Morton, J.P.; Stellingwerff, T.; Maughan, R.J. Toward a Common Understanding of Diet-Exercise Strategies to Manipulate Fuel Availability for Training and Competition Preparation in Endurance Sport. Int. J. Sport Nutr. Exerc. Metab. 2018, 28, 451–463. [Google Scholar] [CrossRef]
- Burke, L.M.; Ross, M.L.; Garvican-Lewis, L.A.; Welvaert, M.; Heikura, I.A.; Forbes, S.G.; Mirtschin, J.G.; Cato, L.E.; Strobel, N.; Sharma, A.P.; et al. Low carbohydrate, high fat diet impairs exercise economy and negates the performance benefit from intensified training in elite race walkers. J. Physiol. 2017, 595, 2785–2807. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, E.R.; Zhong, L.; Talley, N.J.; Morrison, M.; Holtmann, G. Characterisation of the gastrointestinal mucosa-associated microbiota: A novel technique to prevent cross-contamination during endoscopic procedures. Aliment. Pharmacol. Ther. 2016, 43, 1186–1196. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 2017, 33, 782–783. [Google Scholar] [CrossRef] [PubMed]
- Le Cao, K.A.; Costello, M.E.; Lakis, V.A.; Bartolo, F.; Chua, X.Y.; Brazeilles, R.; Rondeau, P. MixMC: A Multivariate Statistical Framework to Gain Insight into Microbial Communities. PLoS ONE 2016, 11, e0160169. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.T.; Erdman, K.A.; Burke, L.M. American College of Sports Medicine Joint Position Statement. Nutrition and Athletic Performance. Med. Sci. Sports Exerc. 2016, 48, 543–568. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507. [Google Scholar] [CrossRef]
- A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. Available online: http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002863 (accessed on 29 November 2017).
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Moore, W.E.; Moore, L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995, 61, 3202–3207. [Google Scholar]
- Foditsch, C.; Santos, T.M.; Teixeira, A.G.; Pereira, R.V.; Dias, J.M.; Gaeta, N.; Bicalho, R.C. Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS ONE 2014, 9, e116465. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Khan, T.M.; Duncan, S.H.; Harmsen, H.J.; Garcia-Gil, L.J.; Flint, H.J. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids and host-derived substrates for growth. Appl. Environ. Microbiol. 2012, 78, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Baker, S.S.; Nugent, C.A.; Tsompana, M.; Guan, L.; Wang, Y.; Buck, M.J.; Genco, R.J.; Baker, R.D.; Zhu, R.; et al. High-fat diet increases Clostridium clusters XIVa in obese rodents. FASEB J. 2017, 31, 965–969. [Google Scholar]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-fat feeding rather than obesity drives taxonomical and functional changes in the gut microbiota in mice. Microbiome 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Nyman, M.; Fåk, F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol. Nutr. Food Res. 2015, 59, 2066–2076. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Prykhodko, O.; Hållenius, F.F.; Nyman, M. Effects of monobutyrin and tributyrin on liver lipid profile, caecal microbiota composition and SCFA in high-fat diet-fed rats. J. Nutr. Sci. 2017. [Google Scholar] [CrossRef]
- Chen, D.; Yang, Z.; Chen, X.; Huang, Y.; Yin, B.; Guo, F.; Zhao, H.; Huang, J.; Wu, Y.; Gu, R. Effect of Lactobacillus rhamnosus hsryfm 1301 on the Gut Microbiota and Lipid Metabolism in Rats Fed a High-Fat Diet. J. Microbiol. Biotechnol. 2015, 25, 687–695. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Shankar, V.; Gouda, M.; Moncivaiz, J.; Gordon, A.; Reo, N.V.; Hussein, L.; Paliy, O. Differences in Gut Metabolites and Microbial Composition and Functions between Egyptian and U.S. Children Are Consistent with Their Diets. mSystems 2017. [Google Scholar] [CrossRef]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murtaza, N.; Burke, L.M.; Vlahovich, N.; Charlesson, B.; O’ Neill, H.; Ross, M.L.; Campbell, K.L.; Krause, L.; Morrison, M. The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers. Nutrients 2019, 11, 261. https://doi.org/10.3390/nu11020261
Murtaza N, Burke LM, Vlahovich N, Charlesson B, O’ Neill H, Ross ML, Campbell KL, Krause L, Morrison M. The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers. Nutrients. 2019; 11(2):261. https://doi.org/10.3390/nu11020261
Chicago/Turabian StyleMurtaza, Nida, Louise M. Burke, Nicole Vlahovich, Bronwen Charlesson, Hayley O’ Neill, Megan L. Ross, Katrina L. Campbell, Lutz Krause, and Mark Morrison. 2019. "The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers" Nutrients 11, no. 2: 261. https://doi.org/10.3390/nu11020261
APA StyleMurtaza, N., Burke, L. M., Vlahovich, N., Charlesson, B., O’ Neill, H., Ross, M. L., Campbell, K. L., Krause, L., & Morrison, M. (2019). The Effects of Dietary Pattern during Intensified Training on Stool Microbiota of Elite Race Walkers. Nutrients, 11(2), 261. https://doi.org/10.3390/nu11020261