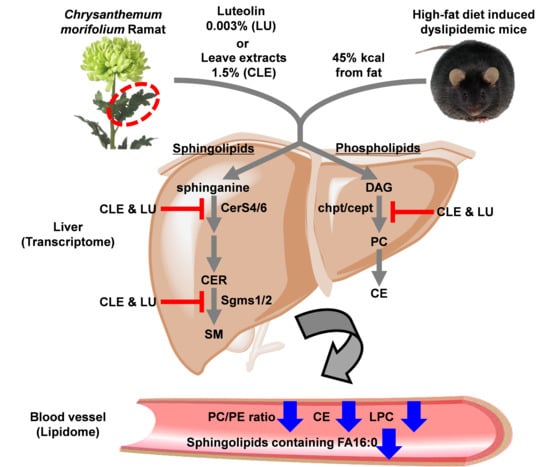
Plasma Lipidomics Reveals Insights into Anti-Obesity Effect of Chrysanthemum morifolium Ramat Leaves and Its Constituent Luteolin in High-Fat Diet-Induced Dyslipidemic Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation of Chrysanthemum morifolium Ramat Leaf Extract
2.3. Animal Experiments
2.4. Lipid Extraction
2.5. Lipidomics Study
2.6. Hepatic Gene Expression Analyses
2.7. Data Processing and Statistical Analyses
3. Results and Discussion
3.1. Animal Characteristics
3.2. Multivariate Statistical Analysis of Mice Plasma Lipid Levels
3.3. Effect of a High-Fat Diet on Mice
3.4. Effect of Chrysanthemum morifolium Ramat leaf Extract (CLE) and Luteolin (LU) in Mice
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Abbreviations | Definition |
| BHT | butylhydroxytoluene |
| CE | cholesteryl ester |
| Cept | ethanolamine phosphotransferase |
| CER | ceramide |
| CerS | ceramide synthase |
| Chpt | choline phosphotransferase |
| CLE | Chrysanthemum morifolium Ramat leaf extracts |
| CM | Chrysanthemum morifolium Ramat |
| CVD | cardiovascular disease |
| DAG | diacylglycerol |
| DES1 | dihydroceramide desaturase |
| HFD | high-fat diet |
| HPLC | high-performance liquid chromatography |
| IS | internal standard |
| kdsr | 3-ketodihydrosphingosine reductase |
| LCAT | lecithin-cholesterol acyltransferase |
| LC-MS/MS | liquid chromatography coupled with tandem mass spectrometry |
| LDL | low-density lipoprotein |
| LPC | lysophosphatidylcholine |
| LPE | lysophosphatidylethanolamine |
| LU | HFD plus luteolin 0.003% diet |
| MTBE | methyl-tert-butyl ether |
| ND | normal diet |
| PC | phosphatidylcholine |
| PCA | principal component analysis |
| PE | phosphatidylethanolamine |
| PEMT | Phosphatidylethanolamine N-methyltransferase |
| PLA2 | phospholipase A2 |
| PLS-DA | partial least squares-discriminant analysis |
| PPAR | peroxisome proliferator-activated receptor |
| QC | quality control |
| Sgms | sphingomyelin synthase |
| SM | sphingomyelin |
| SPT | serine palmitoyltransferase |
| SRM | selected reaction monitoring |
| TAG | triacylglycerol |
| TNF | tumor necrosis factor |
| T2DM | type 2 diabetes mellitus |
| WAT | white adipose tissue |
References
- Cheng, H.; Xu, N.; Zhao, W.; Su, J.; Liang, M.; Xie, Z.; Wu, X.; Li, Q. (-)-Epicatechin regulates blood lipids and attenuates hepatic steatosis in rats fed high-fat diet. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, G.B.D.O.; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef]
- Gadde, K.M.; Martin, C.K.; Berthoud, H.R.; Heymsfield, S.B. Obesity: Pathophysiology and Management. J. Am. Coll. Cardiol. 2018, 71, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Chin, K.Y.; Suhaimi, F.H.; Fairus, A.; Ima-Nirwana, S. Animal models of metabolic syndrome: A review. Nutr. Metab. Lond. 2016, 13, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Y.; Liao, J.K. A mouse model of diet-induced obesity and insulin resistance. Methods Mol. Biol. 2012, 821, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buettner, R.; Scholmerich, J.; Bollheimer, L.C. High-fat diets: Modeling the metabolic disorders of human obesity in rodents. Obesity 2007, 15, 798–808. [Google Scholar] [CrossRef]
- Han, A.R.; Kim, H.Y.; So, Y.; Nam, B.; Lee, I.S.; Nam, J.W.; Jo, Y.D.; Kim, S.H.; Kim, J.B.; Kang, S.Y.; et al. Quantification of Antioxidant Phenolic Compounds in a New Chrysanthemum Cultivar by High-Performance Liquid Chromatography with Diode Array Detection and Electrospray Ionization Mass Spectrometry. Int. J. Anal. Chem. 2017, 2017, 1254721. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, Y.S. Identification of new dicaffeoylquinic acids from Chrysanthemum morifolium and their antioxidant activities. Planta Med. 2005, 71, 871–876. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yang, P.; Luo, Y.; Gao, B.; Sun, J.; Lu, W.; Liu, J.; Chen, P.; Zhang, Y.; Yu, L.L. Chemical compositions of chrysanthemum teas and their anti-inflammatory and antioxidant properties. Food Chem. 2019, 286, 8–16. [Google Scholar] [CrossRef]
- Ukiya, M.; Akihisa, T.; Tokuda, H.; Suzuki, H.; Mukainaka, T.; Ichiishi, E.; Yasukawa, K.; Kasahara, Y.; Nishino, H. Constituents of Compositae plants III. Anti-tumor promoting effects and cytotoxic activity against human cancer cell lines of triterpene diols and triols from edible chrysanthemum flowers. Cancer Lett. 2002, 177, 7–12. [Google Scholar] [CrossRef]
- Gao, T.; Zhu, Z.Y.; Zhou, X.; Xie, M.L. Chrysanthemum morifolium extract improves hypertension-induced cardiac hypertrophy in rats by reduction of blood pressure and inhibition of myocardial hypoxia inducible factor-1alpha expression. Pharm. Biol. 2016, 54, 2895–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Wang, X.; Xue, J.; Liu, J.; Xie, M. Chrysanthemum morifolium extract attenuates high-fat milk-induced fatty liver through peroxisome proliferator-activated receptor alpha-mediated mechanism in mice. Nutr. Res. 2014, 34, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Nepali, S.; Cha, J.Y.; Ki, H.H.; Lee, H.Y.; Kim, Y.H.; Kim, D.K.; Song, B.J.; Lee, Y.M. Chrysanthemum indicum Inhibits Adipogenesis and Activates the AMPK Pathway in High-Fat-Diet-Induced Obese Mice. Am. J. Chin. Med. 2018, 46, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Zhu, Z.-Y.; Wang, F.; Liu, J.-C.; Liu, J.-Y.; Xie, M.-L. Hypoglycemic effect of Chrysanthemum morifolium extract on alloxan-induced diabetic mice is associated with peroxisome proliferator-activated receptor α/γ-mediated hepatic glycogen synthesis. J. Appl. Biomed. 2017, 15, 81–86. [Google Scholar] [CrossRef]
- Lai, J.P.; Lim, Y.H.; Su, J.; Shen, H.M.; Ong, C.N. Identification and characterization of major flavonoids and caffeoylquinic acids in three Compositae plants by LC/DAD-APCI/MS. J. Chromatogr. B Anal. Technol. Biomed Life Sci. 2007, 848, 215–225. [Google Scholar] [CrossRef]
- Wang, T.; Shen, X.G.; Guo, Q.S.; Zhou, J.S.; Mao, P.F.; Shen, Z.G. Comparison of major bioactive components from leaves of Chrysanthemum morifolium. China J. Chin. Mater. Med. 2015, 40, 1670–1675. [Google Scholar]
- Ryu, R.; Kwon, E.Y.; Choi, J.Y.; Shon, J.C.; Liu, K.H.; Choi, M.S. Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue. Nutrients 2019, 11, 1347. [Google Scholar] [CrossRef] [Green Version]
- Huynh, K.; Barlow, C.K.; Jayawardana, K.S.; Weir, J.M.; Mellett, N.A.; Cinel, M.; Magliano, D.J.; Shaw, J.E.; Drew, B.G.; Meikle, P.J. High-Throughput Plasma Lipidomics: Detailed Mapping of the Associations with Cardiometabolic Risk Factors. Cell Chem. Biol. 2019, 26, 71–84. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Baillie, R.A.; Zhu, H.; Zeng, Z.B.; Wiest, M.M.; Nguyen, U.T.; Watkins, S.M.; Krauss, R.M. Lipidomic analysis of variation in response to simvastatin in the Cholesterol and Pharmacogenetics Study. Metabolomics 2010, 6, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.H.; Vaziri, N.D.; Wei, F.; Cheng, X.L.; Bai, X.; Zhao, Y.Y. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci. Rep. 2016, 6, 22151. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishanan, N.; Denna, T.; Devaraj, S.; Adams-Huet, B.; Jialal, I. Exploratory lipidomics in patients with nascent Metabolic Syndrome. J. Diabetes Complicat. 2018, 32, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Shah, C.; Yang, G.; Lee, I.; Bielawski, J.; Hannun, Y.A.; Samad, F. Protection from high fat diet-induced increase in ceramide in mice lacking plasminogen activator inhibitor 1. J. Biol. Chem. 2008, 283, 13538–13548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisinger, K.; Liebisch, G.; Schmitz, G.; Aslanidis, C.; Krautbauer, S.; Buechler, C. Lipidomic analysis of serum from high fat diet induced obese mice. Int. J. Mol. Sci. 2014, 15, 2991–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, L.; Ning, Z.W.; Huang, T.; Wen, B.; Liao, C.H.; Lin, C.Y.; Zhao, L.; Xiao, H.T.; Bian, Z.X. Cyclocarya paliurus Leaves Tea Improves Dyslipidemia in Diabetic Mice: A Lipidomics-Based Network Pharmacology Study. Front. Pharmacol. 2018, 9, 973. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.; Choi, M.S.; Choi, J.Y.; Kim, N.; Kim, M.S.; Jung, S.; Kim, J.; Ryu, D.H.; Hwang, G.S. Effect of green tea on hepatic lipid metabolism in mice fed a high-fat diet. J. Nutr. Biochem. 2018, 51, 1–7. [Google Scholar] [CrossRef]
- Shon, J.C.; Shin, H.S.; Seo, Y.K.; Yoon, Y.R.; Shin, H.; Liu, K.H. Direct infusion MS-based lipid profiling reveals the pharmacological effects of compound K-reinforced ginsenosides in high-fat diet induced obese mice. J. Agric. Food Chem. 2015, 63, 2919–2929. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liang, Y.; Lu, J. Effect of different extracting methods on quality of Chrysanthemum Morifolium Ramat. Infusion. Asia Pac. J. Clin. Nutr. 2007, 16 (Suppl. 1), 183–187. [Google Scholar]
- Duan, K.; Yuan, Z.; Guo, W.; Meng, Y.; Cui, Y.; Kong, D.; Zhang, L.; Wang, N. LC-MS/MS determination and pharmacokinetic study of five flavone components after solvent extraction/acid hydrolysis in rat plasma after oral administration of Verbena officinalis L. extract. J. Ethnopharmacol. 2011, 135, 201–208. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Harnly, J.M. Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem. 2010, 120, 319–326. [Google Scholar] [CrossRef]
- Tsimogiannis, D.; Samiotaki, M.; Panayotou, G.; Oreopoulou, V. Characterization of flavonoid subgroups and hydroxy substitution by HPLC-MS/MS. Molecules 2007, 12, 593–606. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shon, J.C.; Noh, Y.J.; Kwon, Y.S.; Kim, J.H.; Wu, Z.; Seo, J.S. The impact of phenanthrene on membrane phospholipids and its biodegradation by Sphingopyxis soli. Ecotoxicol. Environ. Saf. 2020, 192, 110254. [Google Scholar] [CrossRef] [PubMed]
- Im, S.S.; Park, H.Y.; Shon, J.C.; Chung, I.S.; Cho, H.C.; Liu, K.H.; Song, D.K. Plasma sphingomyelins increase in pre-diabetic Korean men with abdominal obesity. PLoS ONE 2019, 14, e0213285. [Google Scholar] [CrossRef] [Green Version]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cajka, T.; Smilowitz, J.T.; Fiehn, O. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem. 2017, 89, 12360–12368. [Google Scholar] [CrossRef] [PubMed]
- Karsai, G.; Kraft, F.; Haag, N.; Korenke, G.C.; Hanisch, B.; Othman, A.; Suriyanarayanan, S.; Steiner, R.; Knopp, C.; Mull, M.; et al. DEGS1-associated aberrant sphingolipid metabolism impairs nervous system function in humans. J. Clin. Investig. 2019, 129, 1229–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Burla, B.; Arita, M.; Arita, M.; Bendt, A.K.; Cazenave-Gassiot, A.; Dennis, E.A.; Ekroos, K.; Han, X.; Ikeda, K.; Liebisch, G.; et al. MS-based lipidomics of human blood plasma: A community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018, 59, 2001–2017. [Google Scholar] [CrossRef] [Green Version]
- Lange, M.; Fedorova, M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST(R) SRM(R) 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Zacek, P.; Bukowski, M.; Mehus, A.; Johnson, L.; Zeng, H.; Raatz, S.; Idso, J.P.; Picklo, M. Dietary saturated fatty acid type impacts obesity-induced metabolic dysfunction and plasma lipidomic signatures in mice. J. Nutr. Biochem. 2019, 64, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, J.N.; Choi, J.H.; Cha, Y.S.; Muthaiya, M.J.; Lee, C.H. Effect of fermented soybean product (Cheonggukjang) intake on metabolic parameters in mice fed a high-fat diet. Mol. Nutr. Food Res. 2013, 57, 1886–1891. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J.H.; Noh, S.; Hur, H.J.; Sung, M.J.; Hwang, J.T.; Park, J.H.; Yang, H.J.; Kim, M.S.; Kwon, D.Y.; et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Proteome Res. 2011, 10, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Petkevicius, K.; Virtue, S.; Bidault, G.; Jenkins, B.; Cubuk, C.; Morgantini, C.; Aouadi, M.; Dopazo, J.; Serlie, M.J.; Koulman, A.; et al. Accelerated phosphatidylcholine turnover in macrophages promotes adipose tissue inflammation in obesity. Elife 2019, 8. [Google Scholar] [CrossRef]
- Jacobs, R.L.; Zhao, Y.; Koonen, D.P.; Sletten, T.; Su, B.; Lingrell, S.; Cao, G.; Peake, D.A.; Kuo, M.S.; Proctor, S.D.; et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010, 285, 22403–22413. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Su, B.; Jacobs, R.L.; Kennedy, B.; Francis, G.A.; Waddington, E.; Brosnan, J.T.; Vance, J.E.; Vance, D.E. Lack of phosphatidylethanolamine N-methyltransferase alters plasma VLDL phospholipids and attenuates atherosclerosis in mice. Arter. Thromb. Vasc. Biol. 2009, 29, 1349–1355. [Google Scholar] [CrossRef] [Green Version]
- Funai, K.; Lodhi, I.J.; Spears, L.D.; Yin, L.; Song, H.; Klein, S.; Semenkovich, C.F. Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 2016, 65, 358–370. [Google Scholar] [CrossRef] [Green Version]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef]
- Schmitz, G.; Ruebsaamen, K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis 2010, 208, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Law, S.H.; Chan, M.L.; Marathe, G.K.; Parveen, F.; Chen, C.H.; Ke, L.Y. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int. J. Mol. Sci. 2019, 20, 1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegemann, C.; Pechlaner, R.; Willeit, P.; Langley, S.R.; Mangino, M.; Mayr, U.; Menni, C.; Moayyeri, A.; Santer, P.; Rungger, G.; et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014, 129, 1821–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshehry, Z.H.; Mundra, P.A.; Barlow, C.K.; Mellett, N.A.; Wong, G.; McConville, M.J.; Simes, J.; Tonkin, A.M.; Sullivan, D.R.; Barnes, E.H.; et al. Plasma Lipidomic Profiles Improve on Traditional Risk Factors for the Prediction of Cardiovascular Events in Type 2 Diabetes Mellitus. Circulation 2016, 134, 1637–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammad, S.M.; Pierce, J.S.; Soodavar, F.; Smith, K.J.; Al Gadban, M.M.; Rembiesa, B.; Klein, R.L.; Hannun, Y.A.; Bielawski, J.; Bielawska, A. Blood sphingolipidomics in healthy humans: Impact of sample collection methodology. J. Lipid Res. 2010, 51, 3074–3087. [Google Scholar] [CrossRef] [Green Version]
- Samad, F.; Hester, K.D.; Yang, G.; Hannun, Y.A.; Bielawski, J. Altered adipose and plasma sphingolipid metabolism in obesity: A potential mechanism for cardiovascular and metabolic risk. Diabetes 2006, 55, 2579–2587. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Badeanlou, L.; Bielawski, J.; Roberts, A.J.; Hannun, Y.A.; Samad, F. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E211–E224. [Google Scholar] [CrossRef] [Green Version]
- Kasumov, T.; Li, L.; Li, M.; Gulshan, K.; Kirwan, J.P.; Liu, X.; Previs, S.; Willard, B.; Smith, J.D.; McCullough, A. Ceramide as a mediator of non-alcoholic Fatty liver disease and associated atherosclerosis. PLoS ONE 2015, 10, e0126910. [Google Scholar] [CrossRef]
- Adams, J.M., 2nd; Pratipanawatr, T.; Berria, R.; Wang, E.; DeFronzo, R.A.; Sullards, M.C.; Mandarino, L.J. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes 2004, 53, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Cowart, L.A. Sphingolipids and Metabolic Disease; Springer Science & Business Media: Berlin, Germany, 2011; Volume 721. [Google Scholar]
- Longato, L.; Tong, M.; Wands, J.R.; de la Monte, S.M. High fat diet induced hepatic steatosis and insulin resistance: Role of dysregulated ceramide metabolism. Hepatol. Res. 2012, 42, 412–427. [Google Scholar] [CrossRef] [Green Version]
- Cinar, R.; Godlewski, G.; Liu, J.; Tam, J.; Jourdan, T.; Mukhopadhyay, B.; Harvey-White, J.; Kunos, G. Hepatic cannabinoid-1 receptors mediate diet-induced insulin resistance by increasing de novo synthesis of long-chain ceramides. Hepatology 2014, 59, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turpin, S.M.; Nicholls, H.T.; Willmes, D.M.; Mourier, A.; Brodesser, S.; Wunderlich, C.M.; Mauer, J.; Xu, E.; Hammerschmidt, P.; Bronneke, H.S.; et al. Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab. 2014, 20, 678–686. [Google Scholar] [CrossRef] [Green Version]
- Raichur, S.; Brunner, B.; Bielohuby, M.; Hansen, G.; Pfenninger, A.; Wang, B.; Bruning, J.C.; Larsen, P.J.; Tennagels, N. The role of C16:0 ceramide in the development of obesity and type 2 diabetes: CerS6 inhibition as a novel therapeutic approach. Mol. Metab. 2019, 21, 36–50. [Google Scholar] [CrossRef]
- Raichur, S.; Wang, S.T.; Chan, P.W.; Li, Y.; Ching, J.; Chaurasia, B.; Dogra, S.; Ohman, M.K.; Takeda, K.; Sugii, S.; et al. CerS2 haploinsufficiency inhibits beta-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab. 2014, 20, 687–695. [Google Scholar] [CrossRef] [Green Version]
- Fucho, R.; Casals, N.; Serra, D.; Herrero, L. Ceramides and mitochondrial fatty acid oxidation in obesity. FASEB J. 2017, 31, 1263–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Fan, Y.; Liu, J.; Li, Y.; Huan, C.; Bui, H.H.; Kuo, M.S.; Park, T.S.; Cao, G.; Jiang, X.C. Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice. Arter. Thromb. Vasc. Biol. 2012, 32, 1577–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsutake, S.; Zama, K.; Yokota, H.; Yoshida, T.; Tanaka, M.; Mitsui, M.; Ikawa, M.; Okabe, M.; Tanaka, Y.; Yamashita, T.; et al. Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, fatty liver, and type 2 diabetes. J. Biol. Chem. 2011, 286, 28544–28555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dong, J.; Ding, T.; Kuo, M.S.; Cao, G.; Jiang, X.C.; Li, Z. Sphingomyelin synthase 2 activity and liver steatosis: An effect of ceramide-mediated peroxisome proliferator-activated receptor gamma2 suppression. Arter. Thromb. Vasc. Biol. 2013, 33, 1513–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shon, J.C.; Kim, W.C.; Ryu, R.; Wu, Z.; Seo, J.-S.; Choi, M.-S.; Liu, K.-H. Plasma Lipidomics Reveals Insights into Anti-Obesity Effect of Chrysanthemum morifolium Ramat Leaves and Its Constituent Luteolin in High-Fat Diet-Induced Dyslipidemic Mice. Nutrients 2020, 12, 2973. https://doi.org/10.3390/nu12102973
Shon JC, Kim WC, Ryu R, Wu Z, Seo J-S, Choi M-S, Liu K-H. Plasma Lipidomics Reveals Insights into Anti-Obesity Effect of Chrysanthemum morifolium Ramat Leaves and Its Constituent Luteolin in High-Fat Diet-Induced Dyslipidemic Mice. Nutrients. 2020; 12(10):2973. https://doi.org/10.3390/nu12102973
Chicago/Turabian StyleShon, Jong Cheol, Won Cheol Kim, Ri Ryu, Zhexue Wu, Jong-Su Seo, Myung-Sook Choi, and Kwang-Hyeon Liu. 2020. "Plasma Lipidomics Reveals Insights into Anti-Obesity Effect of Chrysanthemum morifolium Ramat Leaves and Its Constituent Luteolin in High-Fat Diet-Induced Dyslipidemic Mice" Nutrients 12, no. 10: 2973. https://doi.org/10.3390/nu12102973
APA StyleShon, J. C., Kim, W. C., Ryu, R., Wu, Z., Seo, J. -S., Choi, M. -S., & Liu, K. -H. (2020). Plasma Lipidomics Reveals Insights into Anti-Obesity Effect of Chrysanthemum morifolium Ramat Leaves and Its Constituent Luteolin in High-Fat Diet-Induced Dyslipidemic Mice. Nutrients, 12(10), 2973. https://doi.org/10.3390/nu12102973