Curcumin from Turmeric Rhizome: A Potential Modulator of DNA Methylation Machinery in Breast Cancer Inhibition
Abstract
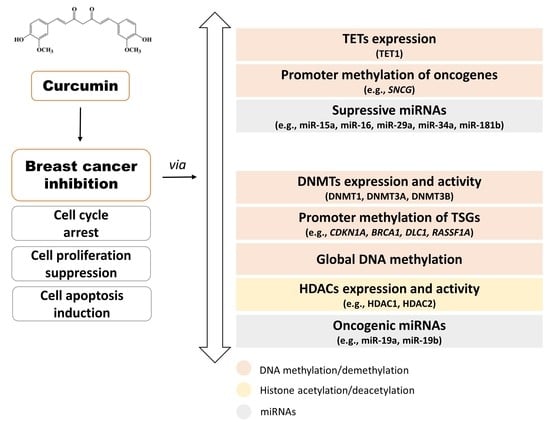
:1. Introduction
2. Curcumin: Chemical Structure and Physical Properties
3. DNA Methylation and Demethylation Processes
4. Curcumin as an Epigenetic Inhibitor of Mammary Cancer
4.1. Curcumin and DNMTs
4.2. Curcumin and HDACs/HATs
4.3. Curcumin and miRNAs
4.4. Curcumin Epigenetic Anti-Cancer Effects Revealed in In Vivo Studies
4.5. Curcumin Epigenetic Anti-Cancer Effects Revealed in Clinical Trials
5. Insight into the Other Bioactive Components of Turmeric Rhizome as Potential Epigenetic Modifiers
6. Discussion, Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2020. [Google Scholar] [CrossRef] [Green Version]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [Green Version]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moo, T.A.; Sanford, R.; Dang, C.; Morrow, M. Overview of breast cancer therapy. PET Clin. 2018, 13, 339–354. [Google Scholar] [CrossRef]
- Singhal, S.K.; Usmani, N.; Michiels, S.; Metzger-Filho, O.; Saini, K.S.; Kovalchuk, O.; Parliament, M. Towards understanding the breast cancer epigenome: A comparison of genome-wide DNA methylation and gene expression data. Oncotarget 2016, 7, 3002–3017. [Google Scholar] [CrossRef] [Green Version]
- Young, G.P.; Le Leu, R.K. Preventing cancer: Dietary lifestyle or clinical intervention? Preventing cancer: Diet or screening? Asia Pac. J. Clin. Nutr. 2002, 11, S618–S631. [Google Scholar] [CrossRef] [Green Version]
- Chlebowski, R.T.; Chen, Z.; Anderson, G.L.; Rohan, T.; Aragaki, A.; Lane, D.; Dolan, N.C.; Paskett, E.D.; McTiernan, A.; Hubbell, F.A.; et al. Ethnicity and breast cancer: Factors influencing differences in incidence and outcome. JNCI J. Natl. Cancer Inst. 2005, 97, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Youlden, D.R.; Cramb, S.M.; Yip, C.H.; Baade, P.D. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol. Med. 2014, 11, 101–115. [Google Scholar] [CrossRef] [Green Version]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A review of its’ effects on human health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Dandawate, P.R.; Subramaniam, D.; Jensen, R.A.; Anant, S. Targeting cancer stem cells and signaling pathways by phytochemicals: Novel approach for breast cancer therapy. Semin. Cancer Biol. 2016, 40–41, 192–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyanapalli, S.S.S.; Kong, A.-N.T. “Curcumin, the king of spices”: Epigenetic regulatory mechanisms in the prevention of cancer, neurological, and inflammatory diseases. Curr. Pharmacol. Rep. 2015, 1, 129–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanska, B.; Karlic, H.; Varga, F.; Fabianowska-Majewska, K.; Haslberger, A. Epigenetic mechanisms in anti-cancer actions of bioactive food components—The implications in cancer prevention. Br. J. Pharmacol. 2012, 167, 279–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banik, U.; Parasuraman, S.; Adhikary, A.K.; Othman, N.H. Curcumin: The spicy modulator of breast carcinogenesis. J. Exp. Clin. Cancer Res. CR 2017, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhavi, D.; Kagan, D. Bioavailability of a sustained release formulation of curcumin. Integr. Med. 2014, 13, 7. [Google Scholar]
- Thorat, B.; Jangle, R. Reversed-phase high-performance liquid chromatography method for analysis of curcuminoids and curcuminoid-loaded liposome formulation. Indian J. Pharm. Sci. 2013, 75, 60. [Google Scholar] [CrossRef] [Green Version]
- Ganiger, S.; Malleshappa, H.N.; Krishnappa, H.; Rajashekhar, G.; Ramakrishna Rao, V.; Sullivan, F. A two generation reproductive toxicity study with curcumin, turmeric yellow, in Wistar rats. Food Chem. Toxicol. 2007, 45, 64–69. [Google Scholar] [CrossRef]
- Joint FAO/WHO Expert Committee on Food Additives. Safety Evaluation of Certain Food Additives and Contaminants; World Health Organization (IPCS): Geneva, Switzerland, 2004; ISBN 978-92-4-166052-5. [Google Scholar]
- FoodData Central. Available online: https://fdc.nal.usda.gov/fdc-app.html#/food-details/172231/nutrients (accessed on 26 November 2020).
- Kurita, T.; Makino, Y. Novel curcumin oral delivery systems. Anticancer Res. 2013, 33, 2807–2821. [Google Scholar]
- Schiborr, C.; Kocher, A.; Behnam, D.; Jandasek, J.; Toelstede, S.; Frank, J. The oral bioavailability of curcumin from micronized powder and liquid micelles is significantly increased in healthy humans and differs between sexes. Mol. Nutr. Food Res. 2014, 58, 516–527. [Google Scholar] [CrossRef]
- Pan, M.-H.; Huang, T.-M.; Lin, J.-K. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab. Dispos. 1999, 27, 486–494. [Google Scholar]
- Frank, J.; Schiborr, C.; Kocher, A.; Meins, J.; Behnam, D.; Schubert-Zsilavecz, M.; Abdel-Tawab, M. Transepithelial transport of curcumin in caco-2 cells is significantly enhanced by micellar solubilisation. Plant Foods Hum. Nutr. 2017, 72, 48–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolat, Z.B.; Islek, Z.; Demir, B.N.; Yilmaz, E.N.; Sahin, F.; Ucisik, M.H. Curcumin- and piperine-loaded emulsomes as combinational treatment approach enhance the anticancer activity of curcumin on HCT116 colorectal cancer model. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thulasidasan, A.K.T.; Retnakumari, A.P.; Shankar, M.; Vijayakurup, V.; Anwar, S.; Thankachan, S.; Pillai, K.S.; Pillai, J.J.; Nandan, C.D.; Alex, V.V.; et al. Folic acid conjugation improves the bioavailability and chemosensitizing efficacy of curcumin-encapsulated PLGA-PEG nanoparticles towards paclitaxel chemotherapy. Oncotarget 2017, 8, 107374–107389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Zhai, Y.; Heng, X.; Che, F.Y.; Chen, W.; Sun, D.; Zhai, G. Oral bioavailability of curcumin: Problems and advancements. J. Drug Target. 2016, 24, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Esteller, M.; Rountree, M.R.; Bachman, K.E.; Schuebel, K.; Herman, J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001, 10, 687–692. [Google Scholar] [CrossRef]
- Das, P.M.; Singal, R. DNA methylation and cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004, 22, 4632–4642. [Google Scholar] [CrossRef] [Green Version]
- Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef] [Green Version]
- Newell-Price, J.; Clark, A.J.; King, P. DNA methylation and silencing of gene expression. Trends Endocrinol. Metab. TEM 2000, 11, 142–148. [Google Scholar] [CrossRef]
- Esteller, M. CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene 2002, 21, 5427–5440. [Google Scholar] [CrossRef] [Green Version]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. CMLS 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Kim, S.J.; Noh, D.Y.; Park, I.A.; Choe, K.J.; Yoo, O.J.; Kang, H.S. Methylation in the p53 promoter is a supplementary route to breast carcinogenesis: Correlation between CpG methylation in the p53 promoter and the mutation of the p53 gene in the progression from ductal carcinoma in situ to invasive ductal carcinoma. Lab. Investig. J. Tech. Methods Pathol. 2001, 81, 573–579. [Google Scholar] [CrossRef] [Green Version]
- Veland, N.; Lu, Y.; Hardikar, S.; Gaddis, S.; Zeng, Y.; Liu, B.; Estecio, M.R.; Takata, Y.; Lin, K.; Tomida, M.W.; et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019, 47, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girault, I.; Tozlu, S.; Lidereau, R.; Bièche, I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003, 9, 4415–4422. [Google Scholar]
- Iida, T.; Suetake, I.; Tajima, S.; Morioka, H.; Ohta, S.; Obuse, C.; Tsurimoto, T. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells Devoted Mol. Cell. Mech. 2002, 7, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.; Ian, H.I.; Koh, T.W.; Ng, H.H.; Xu, G.; Li, B.F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 1997, 277, 1996–2000. [Google Scholar] [CrossRef]
- Milutinovic, S.; Knox, J.D.; Szyf, M. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J. Biol. Chem. 2000, 275, 6353–6359. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Kim, G.-D. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002, 21, 779–788. [Google Scholar] [CrossRef] [Green Version]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef]
- Kimura, H.; Shiota, K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003, 278, 4806–4812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatematsu, K.I.; Yamazaki, T.; Ishikawa, F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells Devoted Mol. Cell. Mech. 2000, 5, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Fuks, F.; Burgers, W.A.; Brehm, A.; Hughes-Davies, L.; Kouzarides, T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000, 24, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Reale, A.; Passananti, C.; Pradhan, S.; Buontempo, S.; De Matteis, G.; Adams, R.L.P.; Caiafa, P. Inhibition of poly(ADP-ribosyl)ation induces DNA hypermethylation: A possible molecular mechanism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1319–1321. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Sarkissyan, M.; Vadgama, J.V. Epigenetics in breast and prostate cancer. Methods Mol. Biol. 2015, 1238, 425–466. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Yang, L.; Yu, S.-J.; Hong, Q.; Yang, Y.; Shao, Z.-M. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs are associated with poor prognosis of patients with early breast cancer. PLoS ONE 2015, 10, e0133896. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Bai, F.; Zhang, J.-Y.; Ma, S.-H.; Liu, J.; Xu, Z.-D.; Zhu, H.-G.; Ling, Z.-Q.; Ye, D.; et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013, 32, 663–669. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Zhou, Y.; Li, Y.; Xu, D.-P.; Li, S.; Li, H.-B. Spices for prevention and treatment of cancers. Nutrients 2016, 8, 495. [Google Scholar] [CrossRef]
- Hu, S.; Xu, Y.; Meng, L.; Huang, L.; Sun, H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp. Ther. Med. 2018, 16, 1266–1272. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, J.; Cui, R.; Lin, J.; Ding, X. Curcumin in treating breast cancer: A review. J. Lab. Autom. 2016, 21, 723–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.-L.; Pan, Y.-Y.; Chen, O.; Luan, Y.; Xue, X.; Zhao, J.-J.; Liu, L.; Jia, H.-Y. Curcumin inhibits MCF-7 cells by modulating the NF-κB signaling pathway. Oncol. Lett. 2017, 14, 5581–5584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunwar, A.; Barik, A.; Mishra, B.; Rathinasamy, K.; Pandey, R.; Priyadarsini, K.I. Quantitative cellular uptake, localization and cytotoxicity of curcumin in normal and tumor cells. Biochimica et Biophysica Acta 2008, 1780, 673–679. [Google Scholar] [CrossRef]
- Ravindran, J.; Prasad, S.; Aggarwal, B.B. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 2009, 11, 495–510. [Google Scholar] [CrossRef]
- Syng-Ai, C.; Kumari, A.L.; Khar, A. Effect of curcumin on normal and tumor cells: Role of glutathione and bcl-2. Mol. Cancer Ther. 2004, 3, 1101–1108. [Google Scholar] [PubMed]
- Abramczyk, H.; Surmacki, J.; Kopeć, M.; Olejnik, A.K.; Lubecka-Pietruszewska, K.; Fabianowska-Majewska, K. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst 2015, 140, 2224–2235. [Google Scholar] [CrossRef] [PubMed]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J. Breast Cancer 2013, 16, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Xiao, Q.; Zhao, L.; Ren, J.; Bai, X.; Sun, M.; Wu, H.; Liu, X.; Song, Z.; Yan, Y.; et al. DNA methyltransferase 1/3a overexpression in sporadic breast cancer is associated with reduced expression of estrogen receptor-alpha/breast cancer susceptibility gene 1 and poor prognosis. Mol. Carcinog. 2015, 54, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharashi, L.A.; Al-Mohanna, F.H.; Tulbah, A.; Aboussekhra, A. The DNA methyl-transferase protein DNMT1 enhances tumor-promoting properties of breast stromal fibroblasts. Oncotarget 2018, 9, 2329–2343. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Jones, W.; Pavlovicz, R.E.; Liu, S.; Yu, J.; Li, P.; Lin, J.; Fuchs, J.R.; Marcucci, G.; et al. Curcumin is a potent DNA hypomethylation agent. Bioorg. Med. Chem. Lett. 2009, 19, 706–709. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Curcumin-mediated demethylation of the proximal promoter CpG island enhances the KLF4 recruitment that leads to increased expression of p21Cip1 in vitro. J. Cell. Biochem. 2019, 120, 809–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Yousef, N.; Shinwari, Z.; Al-Shahrani, B.; Al-Showimi, M.; Al-Moghrabi, N. Curcumin induces re-expression of BRCA1 and suppression of γ synuclein by modulating DNA promoter methylation in breast cancer cell lines. Oncol. Rep. 2020, 43, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhou, J.; Hu, Y.; Wang, J.; Yuan, C. Curcumin inhibits growth of human breast cancer cells through demethylation of DLC1 promoter. Mol. Cell. Biochem. 2017, 425, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Xie, Z.; Wu, L.; Chiu, M.; Lin, J.; Chan, K.K.; Liu, S.; Liu, Z. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr. Cancer 2012, 64, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sarkar, R.; Biswas, J.; Roy, M. Curcumin inhibits histone deacetylase leading to cell cycle arrest and apoptosis via upregulation of p21 in breast cancer cell lines. Int. J. Green Nanotechnol. 2012, 4, 183–197. [Google Scholar] [CrossRef]
- Li, X.; Xie, W.; Xie, C.; Huang, C.; Zhu, J.; Liang, Z.; Deng, F.; Zhu, M.; Zhu, W.; Wu, R.; et al. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7 breast cancer cell proliferation. Phytother. Res. PTR 2014, 28, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, W.; Shi, H.; Xie, X.; Li, L.; Tang, H.; Wu, M.; Kong, Y.; Yang, L.; Gao, J.; et al. Synergistic effects of curcumin with emodin against the proliferation and invasion of breast cancer cells through upregulation of miR-34a. Mol. Cell. Biochem. 2013, 382, 103–111. [Google Scholar] [CrossRef]
- Kronski, E.; Fiori, M.E.; Barbieri, O.; Astigiano, S.; Mirisola, V.; Killian, P.H.; Bruno, A.; Pagani, A.; Rovera, F.; Pfeffer, U.; et al. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and -2. Mol. Oncol. 2014, 8, 581–595. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Cao, Y.; Sun, J.; Zhang, Y. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med. Oncol. 2010, 27, 1114–1118. [Google Scholar] [CrossRef]
- Bora-Tatar, G.; Dayangaç-Erden, D.; Demir, A.S.; Dalkara, S.; Yelekçi, K.; Erdem-Yurter, H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorg. Med. Chem. 2009, 17, 5219–5228. [Google Scholar] [CrossRef]
- Norouzi, S.; Majeed, M.; Pirro, M.; Generali, D.; Sahebkar, A. Curcumin as an adjunct therapy and microRNA modulator in breast cancer. Curr. Pharm. Des. 2018, 24, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Bimonte, S.; Barbieri, A.; Palma, G.; Rea, D.; Luciano, A.; D’Aiuto, M.; Arra, C.; Izzo, F. Dissecting the role of curcumin in tumour growth and angiogenesis in mouse model of human breast cancer. BioMed Res. Int. 2015, 2015, 878134. [Google Scholar] [CrossRef] [PubMed]
- Bachmeier, B.; Nerlich, A.G.; Iancu, C.M.; Cilli, M.; Schleicher, E.; Vené, R.; Dell’Eva, R.; Jochum, M.; Albini, A.; Pfeffer, U. The chemopreventive polyphenol curcumin prevents hematogenous breast cancer metastases in immunodeficient mice. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2007, 19, 137–152. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Wang, X.; Liu, X.; Zhang, H.; Lu, Y.; Su, S. Curcumin enhanced antiproliferative effect of mitomycin C in human breast cancer MCF-7 cells in vitro and in vivo. Acta Pharmacologica Sinica 2011, 32, 1402–1410. [Google Scholar] [CrossRef] [Green Version]
- Bayet-Robert, M.; Kwiatkowski, F.; Leheurteur, M.; Gachon, F.; Planchat, E.; Abrial, C.; Mouret-Reynier, M.-A.; Durando, X.; Barthomeuf, C.; Chollet, P. Phase I dose escalation trial of docetaxel plus curcumin in patients with advanced and metastatic breast cancer. Cancer Biol. Ther. 2010, 9, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Saghatelyan, T.; Tananyan, A.; Janoyan, N.; Tadevosyan, A.; Petrosyan, H.; Hovhannisyan, A.; Hayrapetyan, L.; Arustamyan, M.; Arnhold, J.; Rotmann, A.-R.; et al. Efficacy and safety of curcumin in combination with paclitaxel in patients with advanced, metastatic breast cancer: A comparative, randomized, double-blind, placebo-controlled clinical trial. Phytomedicine 2020, 70, 153218. [Google Scholar] [CrossRef]
- Soda, K. Polyamine metabolism and gene methylation in conjunction with one-carbon metabolism. Int. J. Mol. Sci. 2018, 19, 3106. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tollefsbol, T.O. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr. Med. Chem. 2010, 17, 2141–2151. [Google Scholar] [CrossRef]
- Yi, P.; Melnyk, S.; Pogribna, M.; Pogribny, I.P.; Hine, R.J.; James, S.J. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J. Biol. Chem. 2000, 275, 29318–29323. [Google Scholar] [CrossRef] [Green Version]
- Waterland, R.A. Assessing the effects of high methionine intake on DNA methylation. J. Nutr. 2006, 136, 1706S–1710S. [Google Scholar] [CrossRef]
- James, S.J.; Pogribny, I.P.; Pogribna, M.; Miller, B.J.; Jernigan, S.; Melnyk, S. Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J. Nutr. 2003, 133, 3740S–3747S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.D.; Uthus, E.O. DNA methylation, cancer susceptibility, and nutrient interactions. Exp. Biol. Med. 2004, 229, 988–995. [Google Scholar] [CrossRef]
- Duthie, S.J. Folate and cancer: How DNA damage, repair and methylation impact on colon carcinogenesis. J. Inherit. Metab. Dis. 2011, 34, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 2012, 3, 21–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Huang, P.; Law, S.; Tian, H.; Leung, W.; Xu, C. Preventive effect of curcumin against chemotherapy-induced side-effects. Front. Pharmacol. 2018, 9, 1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruti, S.S.; Ulrich, C.M.; White, E. Folate and one-carbon metabolism nutrients from supplements and diet in relation to breast cancer risk. Am. J. Clin. Nutr. 2009, 89, 624–633. [Google Scholar] [CrossRef] [Green Version]
- Shrubsole, M.J.; Shu, X.O.; Li, H.-L.; Cai, H.; Yang, G.; Gao, Y.-T.; Gao, J.; Zheng, W. Dietary B vitamin and methionine intakes and breast cancer risk among Chinese women. Am. J. Epidemiol. 2011, 173, 1171–1182. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.J.; Ward, R.L. Folate and one-carbon metabolism and its impact on aberrant DNA methylation in cancer. Adv. Genet. 2010, 71, 79–121. [Google Scholar] [CrossRef]
- Menon, V.P.; Sudheer, A.R. Antioxidant and anti-inflammatory properties of curcumin. Adv. Exp. Med. Biol. 2007, 595, 105–125. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Y.; Zhang, P. Protective effects of curcumin and quercetin during benzo (a) pyrene induced lung carcinogenesis in mice. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1736–1743. [Google Scholar]
- Gupta, N.; Verma, K.; Nalla, S.; Kulshreshtha, A.; Lall, R.; Prasad, S. Free radicals as a double-edged sword: The cancer preventive and therapeutic roles of curcumin. Molecules 2020, 25, 5390. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.-S.; Wang, Q.; Sun, D.-D.; Dai, F.; Zhou, B. Design, synthesis, and evaluation of curcumin derivatives as Nrf2 activators and cytoprotectors against oxidative death. Eur. J. Med. Chem. 2017, 134, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Gahlot, S.; Majumdar, S. Oxidative stress induced by curcumin promotes the death of cutaneous T-cell lymphoma (HuT-78) by disrupting the function of several molecular targets. Mol. Cancer Ther. 2012, 11, 1873–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghtaderi, H.; Sepehri, H.; Attari, F. Combination of arabinogalactan and curcumin induces apoptosis in breast cancer cells in vitro and inhibits tumor growth via overexpression of p53 level in vivo. Biomed. Pharmacother. 2017, 88, 582–594. [Google Scholar] [CrossRef]
- Minor, E.A.; Court, B.L.; Young, J.I.; Wang, G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013, 288, 13669–13674. [Google Scholar] [CrossRef] [Green Version]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.-X. Molecular mechanisms underlying the link between diet and DNA methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef] [Green Version]



| Name | Amount (Min-Max) | Unit |
|---|---|---|
| Water | 12.850 | g |
| Energy | 312.000 | kcal |
| Protein | 9.680 | g |
| Total lipid (fat) | 3.250 | g |
| Carbohydrate | 67.140 | g |
| Fiber, total dietary | 22.700 | g |
| Calcium, Ca | 168.000 | mg |
| Iron, Fe | 55.000 | mg |
| Magnesium, Mg | 208.000 | mg |
| Phosphorus, P | 299.000 | mg |
| Potassium, K | 2080.000 | mg |
| Sodium, Na | 27.000 | mg |
| Zinc, Zn | 4.500 | mg |
| Copper, Cu | 1.300 | mg |
| Manganese, Mn | 19.800 | mg |
| Selenium, Se | 6.200 | µg |
| Vitamin C, total ascorbic acid | 0.700 | mg |
| Vitamin B1 (thiamin) | 0.058 | mg |
| Vitamin B2 (riboflavin) | 0.150 | mg |
| Vitamin B3 (niacin) | 1.350 | mg |
| Vitamin B5 (pantothenic acid) | 0.542 | mg |
| Vitamin B6 (pyridoxine) | 0.107 (0.034–0.180) | mg |
| Folate, total | 20.000 | µg |
| Choline, total | 49.200 | mg |
| Betaine, total | 9.700 | mg |
| Vitamin E (alpha-tocopherol) | 4.430 | mg |
| Vitamin K (phylloquinone) | 13.400 | µg |
| Fatty acids, total saturated | 1.838 | g |
| Fatty acids, total monounsaturated | 0.449 | g |
| Fatty acids, total polyunsaturated | 0.756 | g |
| Curcuminoids | 2.000–9.000 | g |
| DNMTs Expression | Change | Model | Curcumin Treatment | Reference |
|---|---|---|---|---|
| mRNA level | ||||
| decrease in all DNMTs (DNMT1, DNMT3A, DNMT3B) | MCF-7 MDA-MB-231 | IC50—10 µM/96 h | Mirza S. et al., J Breast Cancer, 2013 [59] | |
| decrease in all DNMTs (DNMT1, DNMT3A, DNMT3B) | MCF-7 | 2 and 20 µM/12 and 24 h | Chatterjee B. et al., J Cell Biochem, 2019 [63] | |
| decrease in DNMT1 (without changes in DNMT3A, DNMT3B) | MDA-MB-361 MDA-MB-231 MCF-7 | 40 µM/48 h | Liu Y. et al., Mol Cell Biochem, 2017 [65] | |
| decrease in DNMT1 | MCF-7 | 10 and 20 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] | |
| protein level | ||||
| 2-fold decrease in DNMT1 | MCF-7 MDA-MB-231 | IC50—10 µM/96 h | Mirza S. et al., J Breast Cancer, 2013 [59] | |
| decrease in all DNMTs (DNMT1, DNMT3A, DNMT3B) | MCF-7 | 2 and 20 µM/12 and 24 h | Chatterjee B. et al., J Cell Biochem, 2019 [63] | |
| reduction in DNMT1 protein level increase in DNMT3A and DNMT3B protein level | HCC-38 UACC-3199 T47D | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] | |
| decrease in DNMT1 (without changes in DNMT3A, DNMT3B) | MDA-MB-361 MDA-MB-231 | 40 µM/48 h | Liu Y. et al., Mol Cell Biochem, 2017 [65] | |
| decrease in DNMT1 | MCF-7 | 10 and 20 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] | |
| Other proteins | ||||
| increase in TET1 mRNA and TET1 protein level | HCC-38 | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] | |
| 3-fold decrease in HDAC1 protein level | MCF-7 MDA-MB-231 | IC50—10 µM/96 h | Mirza S. et al., J Breast Cancer, 2013 [59] | |
| decrease in HDAC1 and HDAC2 protein level | MCF-7 MDA-MB-231 | 50 µM/24 h | Mukherjeea S. et al., Int. J. Green Nanotechnol, 2012 [67] | |
| oncogene | decrease in SNCG mRNA (down to 2-fold) and SNCG protein level | T47D HCC-38 | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] |
| tumor suppressor | induction of DLC1 expression on mRNA and protein level | MDA-MB-361 | 20 and 40 µM/48 h | Liu Y. et al., Mol Cell Biochem, 2017 [65] |
| tumor suppressor | increase in BRCA1 mRNA level up to 2-fold with consequent high increase in BRCA1 protein level | HCC-38 UACC-3199 | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] |
| tumor suppressor | increased level of CDKN1A (p21, 2-fold in MDA-MB-231 and 4-fold in MCF-7) | MCF-7 MDA-MB-231 | IC50—10 µM/96 h | Mirza S. et al., J Breast Cancer, 2013 [59] |
| tumor suppressor | increased level of CDKN1A (p21) | MCF-7 MDA-MB-231 | 50 µM/24 h | Mukherjeea S. et al., Int. J. Green Nanotechnol, 2012 [67] |
| tumor suppressor | increased expression of TP53 and KLF4 on mRNA and protein levels | MCF-7 | 2 and 20 µM/12 and 24 h | Chatterjee B. et al., J Cell Biochem, 2019 [63] |
| tumor suppressor | enhanced mRNA and the protein levels of RASSF1A | MCF-7 MDA-MB-231 | 10 and 20 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] |
| transcription factor | reduction in SP1 expression | MDA-MB-361 | 40 µM/48 h | Liu Y. et al., Mol Cell Biochem, 2017 [65] |
| DNMTs activity | ||||
| methylation activity of DNMT1 in nuclear extract decreased by about 70% (compared to the control) | MCF-7 | 10 and 20 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] | |
| Promoter methylation | ||||
| demethylation of the proximal promoter of CDKN1A (p21) | MCF-7 | 2 and 20 µM/12 and 24 h | Chatterjee B. et al., J Cell Biochem, 2019 [63] | |
| hypermethylation of the SNCG promoter | T47D | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] | |
| partial hypomethylation of the BRCA1 promoter | HCC-38 UACC-3199 | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] | |
| demethylation of DLC1 promoter | MDA-MB-361 | 20 and 40 µM/48 h | Liu Y. et al., Mol Cell Biochem, 2017 [65] | |
| decrease in RASSF1A promoter methylation | MCF-7 | 10 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] | |
| Global DNA methylation | ||||
| hypomethylation | MCF-7 | 2 and 20 µM/12 and 24 h | Chatterjee B. et al., J Cell Biochem, 2019 [63] | |
| the global DNA methylation (GDM) decreased by about 30–35% | MCF-7 | 10 µM/72 h | Du L. et al., Nutr Cancer, 2012 [66] | |
| miRNA | ||||
| downregulation of oncogenic miR-19 (modulates downstream proteins: PTEN, AKT1, MDM2, TP53) | MCF-7 | 1 µM/4 days | Li X. et al., Phytother Res, 2014 [68] | |
| upregulation of miR-29b | T47D | 5 and 10 µM/6 days | Al-Yousef N. et al., Oncol Rep, 2020 [64] | |
| upregulation of miR-34a (reduction in BCL2 and BMI1 expression) | MDA-MB-231 MDA-MB-435 | 30 or 34 μM/24 h | Guo J. et al., Mol Cell Biochem, 2013 [69] | |
| upregulation of miR181b (reduction in CXCL1, CXCL2, MMPs expression) | MDA-MB-231 | 25 μM/24 h | Kronski E. et al., Mol Oncol, 2014 [70] | |
| upregulation of miR-15a and miR-16 (reduction in BCL2 expression) | MCF-7 | 10–60 μM/24 h | Yang J. et al., Med Oncol, 2010 [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabianowska-Majewska, K.; Kaufman-Szymczyk, A.; Szymanska-Kolba, A.; Jakubik, J.; Majewski, G.; Lubecka, K. Curcumin from Turmeric Rhizome: A Potential Modulator of DNA Methylation Machinery in Breast Cancer Inhibition. Nutrients 2021, 13, 332. https://doi.org/10.3390/nu13020332
Fabianowska-Majewska K, Kaufman-Szymczyk A, Szymanska-Kolba A, Jakubik J, Majewski G, Lubecka K. Curcumin from Turmeric Rhizome: A Potential Modulator of DNA Methylation Machinery in Breast Cancer Inhibition. Nutrients. 2021; 13(2):332. https://doi.org/10.3390/nu13020332
Chicago/Turabian StyleFabianowska-Majewska, Krystyna, Agnieszka Kaufman-Szymczyk, Aldona Szymanska-Kolba, Jagoda Jakubik, Grzegorz Majewski, and Katarzyna Lubecka. 2021. "Curcumin from Turmeric Rhizome: A Potential Modulator of DNA Methylation Machinery in Breast Cancer Inhibition" Nutrients 13, no. 2: 332. https://doi.org/10.3390/nu13020332
APA StyleFabianowska-Majewska, K., Kaufman-Szymczyk, A., Szymanska-Kolba, A., Jakubik, J., Majewski, G., & Lubecka, K. (2021). Curcumin from Turmeric Rhizome: A Potential Modulator of DNA Methylation Machinery in Breast Cancer Inhibition. Nutrients, 13(2), 332. https://doi.org/10.3390/nu13020332