A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation
Abstract
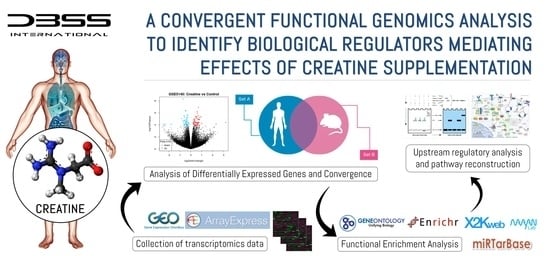
:1. Introduction
2. Methods
2.1. Functional Genomic Analysis
2.1.1. Search and Sources of Evidence
2.1.2. Eligibility Criteria
2.1.3. Analysis of Differentially Expressed Genes and Convergence
2.1.4. Functional Enrichment Analysis
2.1.5. Upstream Regulatory Pathway Analysis
3. Findings
3.1. Selection of Gene Expression Datasets
3.2. Analysis of Differentially Expressed Genes
3.3. Analysis of Convergence between Datasets
3.4. Functional Enrichment Analysis
3.5. Upstream Regulatory Pathway Analysis
4. Discussion
4.1. Biological Pathways Mediating Effects
4.2. Creatine and miRNAs
4.3. The Regulation of the Creatine Transporter
5. Limitations, Strengths, and Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Wallimann, T. The extended, dynamic mitochondrial reticulum in skeletal muscle and the creatine kinase (CK)/phosphocreatine (PCr) shuttle are working hand in hand for optimal energy provision. J. Muscle Res. Cell Motil. 2015, 36, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wallimann, T.; Wyss, M.; Brdiczka, D.; Nicolay, K.; Eppenberger, H.M. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: The ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 1992, 281, 21–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonilla, D.A.; Kreider, R.B.; Stout, J.R.; Forero, D.A.; Kerksick, C.M.; Roberts, M.D.; Rawson, E.S. Metabolic Basis of Creatine in Health and Disease: A Bioinformatics-Assisted Review. Nutrients 2021, 13, 1238. [Google Scholar] [CrossRef] [PubMed]
- Wallimann, T.; Harris, R. Creatine: A miserable life without it. Amino Acids 2016, 48, 1739–1750. [Google Scholar] [CrossRef]
- Kreider, R.B.; Kalman, D.S.; Antonio, J.; Ziegenfuss, T.N.; Wildman, R.; Collins, R.; Candow, D.G.; Kleiner, S.M.; Almada, A.L.; Lopez, H.L. International Society of Sports Nutrition position stand: Safety and efficacy of creatine supplementation in exercise, sport, and medicine. J. Int. Soc. Sports Nutr. 2017, 14, 18. [Google Scholar] [CrossRef]
- Maughan, R.J.; Burke, L.M.; Dvorak, J.; Larson-Meyer, D.E.; Peeling, P.; Phillips, S.M.; Rawson, E.S.; Walsh, N.P.; Garthe, I.; Geyer, H.; et al. IOC consensus statement: Dietary supplements and the high-performance athlete. Br. J. Sports Med. 2018, 52, 439–455. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Kreider, R.B.; Petro, J.L.; Romance, R.; García-Sillero, M.; Benítez-Porres, J.; Vargas-Molina, S. Creatine Enhances the Effects of Cluster-Set Resistance Training on Lower-Limb Body Composition and Strength in Resistance-Trained Men: A Pilot Study. Nutrients 2021, 13, 2303. [Google Scholar] [CrossRef]
- Antonio, J.; Candow, D.G.; Forbes, S.C.; Gualano, B.; Jagim, A.R.; Kreider, R.B.; Rawson, E.S.; Smith-Ryan, A.E.; VanDusseldorp, T.A.; Willoughby, D.S.; et al. Common questions and misconceptions about creatine supplementation: What does the scientific evidence really show? J. Int. Soc. Sports Nutr. 2021, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Kreider, R.B.; Stout, J.R. Creatine in Health and Disease. Nutrients 2021, 13, 447. [Google Scholar] [CrossRef]
- Wallimann, T.; Tokarska-Schlattner, M.; Schlattner, U. The creatine kinase system and pleiotropic effects of creatine. Amino Acids 2011, 40, 1271–1296. [Google Scholar] [CrossRef] [Green Version]
- Wallimann, T.; Hall, C.H.T.; Colgan, S.P.; Glover, L.E. Creatine Supplementation for Patients with Inflammatory Bowl Diseases: A Scientific Rationale for a Clinical Trial. Nutrients 2021, 13, 1429. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, M. Role of Creatine in the Heart: Health and Disease. Nutrients 2021, 13, 1215. [Google Scholar] [CrossRef] [PubMed]
- Smith-Ryan, A.E.; Cabre, H.E.; Eckerson, J.M.; Candow, D.G. Creatine Supplementation in Women’s Health: A Lifespan Perspective. Nutrients 2021, 13, 877. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.; Hickner, R.C.; Ormsbee, M.J. The Potential Role of Creatine in Vascular Health. Nutrients 2021, 13, 857. [Google Scholar] [CrossRef] [PubMed]
- Bredahl, E.C.; Eckerson, J.M.; Tracy, S.M.; McDonald, T.L.; Drescher, K.M. The Role of Creatine in the Development and Activation of Immune Responses. Nutrients 2021, 13, 751. [Google Scholar] [CrossRef] [PubMed]
- Jagim, A.R.; Kerksick, C.M. Creatine Supplementation in Children and Adolescents. Nutrients 2021, 13, 664. [Google Scholar] [CrossRef]
- Solis, M.Y.; Artioli, G.G.; Gualano, B. Potential of Creatine in Glucose Management and Diabetes. Nutrients 2021, 13, 570. [Google Scholar] [CrossRef]
- Roschel, H.; Gualano, B.; Ostojic, S.M.; Rawson, E.S. Creatine Supplementation and Brain Health. Nutrients 2021, 13, 586. [Google Scholar] [CrossRef]
- Muccini, A.M.; Tran, N.T.; de Guingand, D.L.; Philip, M.; Della Gatta, P.A.; Galinsky, R.; Sherman, L.S.; Kelleher, M.A.; Palmer, K.R.; Berry, M.J.; et al. Creatine Metabolism in Female Reproduction, Pregnancy and Newborn Health. Nutrients 2021, 13, 490. [Google Scholar] [CrossRef]
- Li, J.L.; Wang, X.N.; Fraser, S.F.; Carey, M.F.; Wrigley, T.V.; McKenna, M.J. Effects of fatigue and training on sarcoplasmic reticulum Ca2+ regulation in human skeletal muscle. J. Appl. Physiol. 2002, 92, 912–922. [Google Scholar] [CrossRef] [Green Version]
- Sundberg, C.W.; Hunter, S.K.; Trappe, S.W.; Smith, C.S.; Fitts, R.H. Effects of elevated H+ and Pi on the contractile mechanics of skeletal muscle fibres from young and old men: Implications for muscle fatigue in humans. J. Physiol. 2018, 596, 3993–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, A.J.; Place, N.; Westerblad, H. Molecular Basis for Exercise-Induced Fatigue: The Importance of Strictly Controlled Cellular Ca2+ Handling. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundberg, C.W.; Fitts, R.H. Bioenergetic basis of skeletal muscle fatigue. Curr. Opin. Physiol. 2019, 10, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Mesa, J.L.M.; Ruiz, J.R.; Gonzalez-Gross, M.M.; Gutierrez Sainz, A.; Castillo Garzon, M.J. Oral Creatine Supplementation and Skeletal Muscle Metabolism in Physical Exercise. Sports Med. 2002, 32, 903–944. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Wilkerson, D.P.; Fulford, J. Influence of dietary creatine supplementation on muscle phosphocreatine kinetics during knee-extensor exercise in humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R1078–R1087. [Google Scholar] [CrossRef] [Green Version]
- Rico-Sanz, J. Creatine reduces human muscle PCr and pH decrements and Pi accumulation during low-intensity exercise. J. Appl. Physiol. 2000, 88, 1181–1191. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.G.; López de Viñaspre, P.; González de Suso, J.M.; Moreno, A.; Alonso, J.; Cabañas, M.; Pons, V.; Porta, J.; Arús, C. Efecto de la suplementación oral con monohidrato de creatina en el metabolismo energértico muscular y en la composición corporal de sujetos que practican actividad fÍsica. Revista Chilena de Nutrición 2003, 30, 58–63. [Google Scholar] [CrossRef]
- Pulido, S.M.; Passaquin, A.C.; Leijendekker, W.J.; Challet, C.; Wallimann, T.; Rüegg, U.T. Creatine supplementation improves intracellular Ca2+ handling and survival in mdx skeletal muscle cells. FEBS Lett. 1998, 439, 357–362. [Google Scholar] [CrossRef] [Green Version]
- Gallo, M.; MacLean, I.; Tyreman, N.; Martins, K.J.B.; Syrotuik, D.; Gordon, T.; Putman, C.T. Adaptive responses to creatine loading and exercise in fast-twitch rat skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R1319–R1328. [Google Scholar] [CrossRef] [Green Version]
- Pignatti, C.; D’Adamo, S.; Flamigni, F.; Cetrulllo, S. Molecular Mechanisms Linking Nutrition to Metabolic Homeostasis: An Overview Picture of Current Understanding. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 543–564. [Google Scholar] [CrossRef]
- Hurtado-Carneiro, V.; Pérez-García, A.; Alvarez, E.; Sanz, C. PAS Kinase: A Nutrient and Energy Sensor “Master Key” in the Response to Fasting/Feeding Conditions. Front. Endocrinol. 2020, 11, 999. [Google Scholar] [CrossRef]
- Hespel, P.; Op’t Eijnde, B.; Leemputte, M.V.; Ursø, B.; Greenhaff, P.L.; Labarque, V.; Dymarkowski, S.; Hecke, P.V.; Richter, E.A. Oral creatine supplementation facilitates the rehabilitation of disuse atrophy and alters the expression of muscle myogenic factors in humans. J. Physiol. 2001, 536, 625–633. [Google Scholar] [CrossRef]
- Willoughby, D.S.; Rosene, J.M. Effects of Oral Creatine and Resistance Training on Myogenic Regulatory Factor Expression. Med. Sci. Sports Exerc. 2003, 35, 923–929. [Google Scholar] [CrossRef] [Green Version]
- Deldicque, L.; Louis, M.; Theisen, D.; Nielens, H.; Dehoux, M.L.; Thissen, J.-P.; Rennie, M.J.; Francaux, M. Increased IGF mRNA in Human Skeletal Muscle after Creatine Supplementation. Med. Sci. Sports Exerc. 2005, 37, 731–736. [Google Scholar] [CrossRef] [Green Version]
- Saremi, A.; Gharakhanloo, R.; Sharghi, S.; Gharaati, M.R.; Larijani, B.; Omidfar, K. Effects of oral creatine and resistance training on serum myostatin and GASP-1. Mol. Cell. Endocrinol. 2010, 317, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Louis, M.; Van Beneden, R.; Dehoux, M.; Thissen, J.P.; Francaux, M. Creatine increases IGF-I and myogenic regulatory factor mRNA in C2C12 cells. FEBS Lett. 2004, 557, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Cunha, M.P.; Budni, J.; Ludka, F.K.; Pazini, F.L.; Rosa, J.M.; Oliveira, Á.; Lopes, M.W.; Tasca, C.I.; Leal, R.B.; Rodrigues, A.L.S. Involvement of PI3K/Akt Signaling Pathway and Its Downstream Intracellular Targets in the Antidepressant-Like Effect of Creatine. Mol. Neurobiol. 2015, 53, 2954–2968. [Google Scholar] [CrossRef] [PubMed]
- Marzuca-Nassr, G.N.; Fortes, M.A.S.; Guimarães-Ferreira, L.; Murata, G.M.; Vitzel, K.F.; Vasconcelos, D.A.A.; Bassit, R.A.; Curi, R. Short-term creatine supplementation changes protein metabolism signaling in hindlimb suspension. Braz. J. Med. Biol. Res. 2019, 52. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Pavlopoulou, A.; Bagos, P.G. Methods of Analysis and Meta-Analysis for Identifying Differentially Expressed Genes. In Genet. Epidemiol; Humana Press: New York, NY, USA, 2018; pp. 183–210. [Google Scholar]
- Van Laere, S.; Dirix, L.; Vermeulen, P. Molecular profiles to biology and pathways: A systems biology approach. Chin. J. Cancer 2016, 35, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angione, C. Human Systems Biology and Metabolic Modelling: A Review—From Disease Metabolism to Precision Medicine. BioMed Res. Int. 2019, 2019, 8304260. [Google Scholar] [CrossRef]
- Chen, E.Y.; Xu, H.; Gordonov, S.; Lim, M.P.; Perkins, M.H.; Ma′ayan, A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 2012, 28, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Forero, D.A.; González-Giraldo, Y. Convergent functional genomics of cocaine misuse in humans and animal models. Am. J. Drug Alcohol Abus. 2019, 46, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Monaco, G.; van Dam, S.; Casal Novo Ribeiro, J.L.; Larbi, A.; de Magalhães, J.P. A comparison of human and mouse gene co-expression networks reveals conservation and divergence at the tissue, pathway and disease levels. BMC Evol. Biol. 2015, 15, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Levine, J.L.S.; Avila-Quintero, V.; Bloch, M.; Kaffman, A. Systematic review and meta-analysis: Effects of maternal separation on anxiety-like behavior in rodents. Transl. Psychiatry 2020, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, A.B.; Le-Niculescu, H. Convergent Functional Genomics: What we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology 2009, 35, 355–356. [Google Scholar] [CrossRef]
- Forero, D.A. Functional Genomics of Epileptogenesis in Animal Models and Humans. Cell. Mol. Neurobiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, A.B.; Levey, D.F.; Phalen, P.L.; Le-Niculescu, H.; Dainton, H.D.; Jain, N.; Belanger, E.; James, A.; George, S.; Weber, H.; et al. Understanding and predicting suicidality using a combined genomic and clinical risk assessment approach. Mol. Psychiatry 2015, 20, 1266–1285. [Google Scholar] [CrossRef]
- Bonvicini, C.; Faraone, S.V.; Scassellati, C. Common and specific genes and peripheral biomarkers in children and adults with attention-deficit/hyperactivity disorder. World J. Biol. Psychiatry 2018, 19, 80–100. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.K.; Fang, H.; Whistler, T.; Unger, E.R.; Rajeevan, M.S. Convergent genomic studies identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology 2011, 64, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Falker-Gieske, C.; Mott, A.; Franzenburg, S.; Tetens, J. Multi-species transcriptome meta-analysis of the response to retinoic acid in vertebrates and comparative analysis of the effects of retinol and retinoic acid on gene expression in LMH cells. BMC Genom. 2021, 22, 146. [Google Scholar] [CrossRef]
- Le-Niculescu, H.; Roseberry, K.; Gill, S.S.; Levey, D.F.; Phalen, P.L.; Mullen, J.; Williams, A.; Bhairo, S.; Voegtline, T.; Davis, H.; et al. Precision medicine for mood disorders: Objective assessment, risk prediction, pharmacogenomics, and repurposed drugs. Mol. Psychiatry 2021. [Google Scholar] [CrossRef]
- Lévesque, A.; Gagnon-Carignan, S.; Lachance, S. From low- to high-throughput analysis. Bioanalysis 2016, 8, 135–141. [Google Scholar] [CrossRef]
- Ramsay, D.S.; Woods, S.C. Physiological Regulation: How It Really Works. Cell Metab. 2016, 24, 361–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawson, E.S.; Persky, A.M. Mechanisms of muscular adaptations to creatine supplementation: Review article. Int. SportMed J. 2007, 8, 43–53. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Moreno, Y. Molecular and metabolic insights of creatine supplementation on resistance training. Revista Colombiana de Química 2015, 44, 11–18. [Google Scholar] [CrossRef]
- Farshidfar, F.; Pinder, M.A.; Myrie, S.B. Creatine Supplementation and Skeletal Muscle Metabolism for Building Muscle Mass- Review of the Potential Mechanisms of Action. Curr. Protein Pept. Sci. 2017, 18, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.; Klopstock, T. Creatine for neuroprotection in neurodegenerative disease: End of story? Amino Acids 2016, 48, 1929–1940. [Google Scholar] [CrossRef]
- Rae, C.D.; Bröer, S. Creatine as a booster for human brain function. How might it work? Neurochem. Int. 2015, 89, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Riesberg, L.A.; Weed, S.A.; McDonald, T.L.; Eckerson, J.M.; Drescher, K.M. Beyond muscles: The untapped potential of creatine. Int. Immunopharmacol. 2016, 37, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021. [Google Scholar] [CrossRef]
- Ramasamy, A.; Mondry, A.; Holmes, C.C.; Altman, D.G. Key Issues in Conducting a Meta-Analysis of Gene Expression Microarray Datasets. PLoS Med. 2008, 5, e184. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [Green Version]
- Clarke, D.J.B.; Kuleshov, M.V.; Schilder, B.M.; Torre, D.; Duffy, M.E.; Keenan, A.B.; Lachmann, A.; Feldmann, A.S.; Gundersen, G.W.; Silverstein, M.C.; et al. eXpression2Kinases (X2K) Web: Linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018, 46, W171–W179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hangelbroek, R.; Backx, E.; Kersten, S.; van Duynhoven, J.; Verdijk, L.; van Loon, L.; de Groot, L.; Boekschoten, M. Creatine supplementation during knee immobilization attenuates changes in muscle transcriptome. In Proceedings of the Phenotypes and Prevention: The Interplay of Genes, Life-Style and Gut Environment, Copenhagen, Denmark, 5–8 September 2016; p. 59. [Google Scholar]
- Safdar, A.; Yardley, N.J.; Snow, R.; Melov, S.; Tarnopolsky, M.A. Global and targeted gene expression and protein content in skeletal muscle of young men following short-term creatine monohydrate supplementation. Physiol. Genom. 2008, 32, 219–228. [Google Scholar] [CrossRef]
- Bender, A.; Beckers, J.; Schneider, I.; Holter, S.M.; Haack, T.; Ruthsatz, T.; Vogt-Weisenhorn, D.M.; Becker, L.; Genius, J.; Rujescu, D.; et al. Creatine improves health and survival of mice. Neurobiol. Aging 2008, 29, 1404–1411. [Google Scholar] [CrossRef]
- Zervou, S.; Ray, T.; Sahgal, N.; Sebag-Montefiore, L.; Cross, R.; Medway, D.J.; Ostrowski, P.J.; Neubauer, S.; Lygate, C.A. A role for thioredoxin-interacting protein (Txnip) in cellular creatine homeostasis. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E263–E270. [Google Scholar] [CrossRef] [Green Version]
- Saks, V.; Monge, C.; Guzun, R. Philosophical Basis and Some Historical Aspects of Systems Biology: From Hegel to Noble—Applications for Bioenergetic Research. Int. J. Mol. Sci. 2009, 10, 1161–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertsch, B.; Ogden, C.A.; Sidhu, K.; Le-Niculescu, H.; Kuczenski, R.; Niculescu, A.B. Convergent functional genomics: A Bayesian candidate gene identification approach for complex disorders. Methods 2005, 37, 274–279. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattane, N.; Scassellati, C.; D’Aprile, I.; Riva, M.A.; Pariante, C.M. Convergent Functional Genomics approach to prioritize molecular targets of risk in early life stress-related psychiatric disorders. Brain Behav. Immun. Health 2020, 8. [Google Scholar] [CrossRef]
- Sterling, P. Allostasis: A model of predictive regulation. Physiol. Behav. 2012, 106, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Shellman, E.R.; Burant, C.F.; Schnell, S. Network motifs provide signatures that characterize metabolism. Mol. BioSyst. 2013, 9, 352–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Qian, W.; Xu, X.; Bajpai, A.; Guan, K.; Zhang, Z.; Chen, R.; Flamini, V.; Chen, W. Energy-Mediated Machinery Drives Cellular Mechanical Allostasis. Adv. Mater. 2019, 31, 1900453. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef]
- O’Connor, L.; Gilmour, J.; Bonifer, C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J. Biol. Med. 2016, 89, 513–525. [Google Scholar]
- Oldfield, A.J.; Henriques, T.; Kumar, D.; Burkholder, A.B.; Cinghu, S.; Paulet, D.; Bennett, B.D.; Yang, P.; Scruggs, B.S.; Lavender, C.A.; et al. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat. Commun. 2019, 10, 3072. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef]
- Fitieh, A.; Locke, A.J.; Motamedi, M.; Ismail, I.H. The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability. Int. J. Mol. Sci. 2021, 22, 2976. [Google Scholar] [CrossRef]
- Li, X.; Thyssen, G.; Beliakoff, J.; Sun, Z. The Novel PIAS-like Protein hZimp10 Enhances Smad Transcriptional Activity. J. Biol. Chem. 2006, 281, 23748–23756. [Google Scholar] [CrossRef] [Green Version]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Dalton, S. Myc transcription factors: Key regulators behind establishment and maintenance of pluripotency. Regen. Med. 2010, 5, 947–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.-H.; Kwak, Y.; Woo, Y.; Park, C.; Lee, S.-A.; Lee, H.; Park, S.J.; Suh, Y.; Suh, B.K.; Goo, B.S.; et al. Regulation of the actin cytoskeleton by the Ndel1-Tara complex is critical for cell migration. Sci. Rep. 2016, 6, 31827. [Google Scholar] [CrossRef]
- Anderson, S.; Poudel, K.R.; Roh-Johnson, M.; Brabletz, T.; Yu, M.; Borenstein-Auerbach, N.; Grady, W.N.; Bai, J.; Moens, C.B.; Eisenman, R.N.; et al. MYC-nick promotes cell migration by inducing fascin expression and Cdc42 activation. Proc. Natl. Acad. Sci. USA 2016, 113, E5481–E5490. [Google Scholar] [CrossRef] [Green Version]
- Hurlin, P.J.; Huang, J. The MAX-interacting transcription factor network. Semin. Cancer Biol. 2006, 16, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Volodina Iu, L.; Shtil, A.A. Casein kinase 2, the versatile regulator of cell survival. Mol. Biol. 2012, 46, 423–433. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [Green Version]
- Salizzato, V.; Zanin, S.; Borgo, C.; Lidron, E.; Salvi, M.; Rizzuto, R.; Pallafacchina, G.; Donella-Deana, A. Protein kinase CK2 subunits exert specific and coordinated functions in skeletal muscle differentiation and fusogenic activity. FASEB J. 2019, 33, 10648–10667. [Google Scholar] [CrossRef]
- Zheng, Y.; Qin, H.; Frank, S.J.; Deng, L.; Litchfield, D.W.; Tefferi, A.; Pardanani, A.; Lin, F.T.; Li, J.; Sha, B.; et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood 2011, 118, 156–166. [Google Scholar] [CrossRef] [Green Version]
- Bousoik, E.; Montazeri Aliabadi, H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [Green Version]
- Rotwein, P. Regulation of gene expression by growth hormone. Mol. Cell. Endocrinol. 2020, 507, 110788. [Google Scholar] [CrossRef] [PubMed]
- Reis, H.J.; Rosa, D.V.F.; Guimarães, M.M.; Souza, B.R.; Barros, A.G.A.; Pimenta, F.J.; Souza, R.P.; Torres, K.C.L.; Romano-Silva, M.A. Is DARPP-32 a potential therapeutic target? Expert Opin. Ther. Targets 2007, 11, 1649–1661. [Google Scholar] [CrossRef] [PubMed]
- Engmann, O.; Giralt, A.; Gervasi, N.; Marion-Poll, L.; Gasmi, L.; Filhol, O.; Picciotto, M.R.; Gilligan, D.; Greengard, P.; Nairn, A.C.; et al. DARPP-32 interaction with adducin may mediate rapid environmental effects on striatal neurons. Nat. Commun. 2015, 6, 10099. [Google Scholar] [CrossRef] [Green Version]
- Andres, R.H.; Ducray, A.D.; Huber, A.W.; Pérez-Bouza, A.; Krebs, S.H.; Schlattner, U.; Seiler, R.W.; Wallimann, T.; Widmer, H.R. Effects of creatine treatment on survival and differentiation of GABA-ergic neurons in cultured striatal tissue. J. Neurochem. 2005, 95, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.W.; Hwang, S.S.; Kim, H.S.; Lee, K.O.; Kim, M.K.; Lee, W.; Kim, K.; Lee, G.R. Casein kinase 2 is a critical determinant of the balance of Th17 and Treg cell differentiation. Exp. Mol. Med. 2017, 49, e375. [Google Scholar] [CrossRef] [Green Version]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Berindan-Neagoe, I. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef] [Green Version]
- Dhanasekaran, D.N.; Kashef, K.; Lee, C.M.; Xu, H.; Reddy, E.P. Scaffold proteins of MAP-kinase modules. Oncogene 2007, 26, 3185–3202. [Google Scholar] [CrossRef] [Green Version]
- Krishna, M.; Narang, H. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell. Mol. Life Sci. 2008, 65, 3525–3544. [Google Scholar] [CrossRef]
- Gyoeva, F.K. The role of motor proteins in signal propagation. Biochemistry 2014, 79, 849–855. [Google Scholar] [CrossRef]
- Hoffman, L.; Jensen, C.C.; Yoshigi, M.; Beckerle, M.; Weaver, V.M. Mechanical signals activate p38 MAPK pathway-dependent reinforcement of actin via mechanosensitive HspB1. Mol. Biol. Cell 2017, 28, 2661–2675. [Google Scholar] [CrossRef]
- Munnik, T.; Ligterink, W.; Meskiene, I.I.; Calderini, O.; Beyerly, J.; Musgrave, A.; Hirt, H. Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 1999, 20, 381–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, M.J.; Dreyer, H.C.; Fry, C.S.; Glynn, E.L.; Rasmussen, B.B. Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling. J. Appl. Physiol. 2009, 106, 1374–1384. [Google Scholar] [CrossRef] [Green Version]
- Schedel, J.M.; Tanaka, H.; Kiyonaga, A.; Shindo, M.; Schutz, Y. Acute creatine loading enhances human growth hormone secretion. J. Sports Med. Phys. Fit. 2000, 40, 336–342. [Google Scholar]
- Poprzecki, S.; Zając, A.; Czuba, M.; Waskiewicz, Z. The Effects of Terminating Creatine Supplementation and Resistance Training on Anaerobic Power and Chosen Biochemical Variables in Male Subjects. J. Hum. Kinet. 2008, 20, 99–110. [Google Scholar] [CrossRef]
- Burke, D.G.; Candow, D.G.; Chilibeck, P.D.; MacNeil, L.G.; Roy, B.D.; Tarnopolsky, M.A.; Ziegenfuss, T. Effect of creatine supplementation and resistance-exercise training on muscle insulin-like growth factor in young adults. Int. J. Sport Nutr. Exerc. Metab. 2008, 18, 389–398. [Google Scholar] [CrossRef] [Green Version]
- Snow, R.; Wright, C.; Quick, M.; Garnham, A.; Watt, K.; Russell, A. The effects of acute exercise and creatine supplementation on Akt signalling in human skeletal muscle. Mol. Cell 2004, 14, 395–403. [Google Scholar]
- Al-Khalili, L.; Kramer, D.; Wretenberg, P.; Krook, A. Human skeletal muscle cell differentiation is associated with changes in myogenic markers and enhanced insulin-mediated MAPK and PKB phosphorylation. Acta Physiol. Scand. 2004, 180, 395–403. [Google Scholar] [CrossRef]
- Deldicque, L.; Theisen, D.; Bertrand, L.; Hespel, P.; Hue, L.; Francaux, M. Creatine enhances differentiation of myogenic C2C12 cells by activating both p38 and Akt/PKB pathways. Am. J. Physiol. Cell Physiol. 2007, 293, C1263–C1271. [Google Scholar] [CrossRef] [Green Version]
- Deldicque, L.; Atherton, P.; Patel, R.; Theisen, D.; Nielens, H.; Rennie, M.J.; Francaux, M. Effects of resistance exercise with and without creatine supplementation on gene expression and cell signaling in human skeletal muscle. J. Appl. Physiol. 2008, 104, 371–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pazini, F.L.; Cunha, M.P.; Rosa, J.M.; Colla, A.R.; Lieberknecht, V.; Oliveira, Á.; Rodrigues, A.L. Creatine, similar to ketamine, counteracts depressive-like behavior induced by corticosterone via PI3K/Akt/mTOR pathway. Mol Neurobiol. 2016, 53, 6818–6834. [Google Scholar] [CrossRef]
- Ju, J.S.; Smith, J.L.; Oppelt, P.J.; Fisher, J.S. Creatine feeding increases GLUT4 expression in rat skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E347–E352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willoughby, D.S.; Rosene, J. Effects of oral creatine and resistance training on myosin heavy chain expression. Med. Sci. Sports Exerc. 2001, 33, 1674–1681. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, Z.; Yan, H.; Wang, W.; Wu, Z.; Zhang, F.; Zhang, Q.; Shi, G.; Du, J.; Cai, H.; et al. Creatine promotes cancer metastasis through activation of Smad2/3. Cell Metab. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.T.d.S.; Dörr, F.A.; Pinto, E.; Solis, M.Y.; Artioli, G.G.; Fernandes, A.L.; Murai, I.H.; Dantas, W.S.; Seguro, A.C.; Santinho, M.A.R.; et al. Can creatine supplementation form carcinogenic heterocyclic amines in humans? J. Physiol. 2015, 593, 3959–3971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazak, L.; Cohen, P. Creatine metabolism: Energy homeostasis, immunity and cancer biology. Nat. Rev. Endocrinol. 2020, 16, 421–436. [Google Scholar] [CrossRef]
- Patra, S.; Bera, S.; SinhaRoy, S.; Ghoshal, S.; Ray, S.; Basu, A.; Schlattner, U.; Wallimann, T.; Ray, M. Progressive decrease of phosphocreatine, creatine and creatine kinase in skeletal muscle upon transformation to sarcoma. FEBS J. 2008, 275, 3236–3247. [Google Scholar] [CrossRef]
- Pal, A.; Roy, A.; Ray, M. Creatine supplementation with methylglyoxal: A potent therapy for cancer in experimental models. Amino Acids 2016, 48, 2003–2013. [Google Scholar] [CrossRef]
- Storey, K.B.; Hochachka, P.W. Activation of muscle glycolysis: A role for creatine phosphate in phosphofructokinase regulation. FEBS Lett. 1974, 46, 337–339. [Google Scholar] [CrossRef] [Green Version]
- Kemp, R.G. Inhibition of muscle pyruvate kinase by creatine phosphate. J. Biol. Chem. 1973, 248, 3963–3967. [Google Scholar] [CrossRef]
- Bergnes, G.; Yuan, W.; Khandekar, V.S.; O’Keefe, M.M.; Martin, K.J.; Teicher, B.A.; Kaddurah-Daouk, R. Creatine and phosphocreatine analogs: Anticancer activity and enzymatic analysis. Oncol. Res. 1996, 8, 121–130. [Google Scholar]
- Campos-Ferraz, P.L.; Gualano, B.; das Neves, W.; Andrade, I.T.; Hangai, I.; Pereira, R.T.; Bezerra, R.N.; Deminice, R.; Seelaender, M.; Lancha, A.H. Exploratory studies of the potential anti-cancer effects of creatine. Amino Acids 2016, 48, 1993–2001. [Google Scholar] [CrossRef] [PubMed]
- Di Biase, S.; Ma, X.; Wang, X.; Yu, J.; Wang, Y.C.; Smith, D.J.; Zhou, Y.; Li, Z.; Kim, Y.J.; Clarke, N.; et al. Creatine uptake regulates CD8 T cell antitumor immunity. J. Exp. Med. 2019, 216, 2869–2882. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Yang, L. Creatine in T Cell Antitumor Immunity and Cancer Immunotherapy. Nutrients 2021, 13, 1633. [Google Scholar] [CrossRef] [PubMed]
- Maguire, O.A.; Ackerman, S.E.; Szwed, S.K.; Maganti, A.V.; Marchildon, F.; Huang, X.; Kramer, D.J.; Rosas-Villegas, A.; Gelfer, R.G.; Turner, L.E.; et al. Creatine-mediated crosstalk between adipocytes and cancer cells regulates obesity-driven breast cancer. Cell Metab. 2021, 33, 499–512.e496. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Z.; Gao, L.; Qiao, Z.; Yu, M.; Yu, B.; Yang, T. miR-1-3p suppresses proliferation of hepatocellular carcinoma through targeting SOX9. Onco Targets Ther. 2019, 12, 2149–2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, J.; Zhang, B.H.; Li, Y.Y.; Zhong, M.; Ma, W.; Xue, H.; Wen, Y.D.; Cai, Y.D. MiR-1-3p suppresses cell proliferation and invasion and targets STC2 in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8870–8877. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, X.; Jing, L.; Li, Y. MiR-1-3p Inhibits Lung Adenocarcinoma Cell Tumorigenesis via Targeting Protein Regulator of Cytokinesis 1. Front. Oncol. 2019, 9, 120. [Google Scholar] [CrossRef] [Green Version]
- Meyer, S.U.; Thirion, C.; Polesskaya, A.; Bauersachs, S.; Kaiser, S.; Krause, S.; Pfaffl, M.W. TNF-α and IGF1 modify the microRNA signature in skeletal muscle cell differentiation. Cell Commun. Signal. 2015, 13, 4. [Google Scholar] [CrossRef] [Green Version]
- Pelka, K.; Klicka, K.; Grzywa, T.M.; Gondek, A.; Marczewska, J.M.; Garbicz, F.; Szczepaniak, K.; Paskal, W.; Wlodarski, P.K. miR-96-5p, miR-134-5p, miR-181b-5p and miR-200b-3p heterogenous expression in sites of prostate cancer versus benign prostate hyperplasia-archival samples study. Histochem. Cell Biol. 2020. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, L.; Wang, S.; Hong, Q. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem. Biophys. Res. Commun. 2020, 522, 388–394. [Google Scholar] [CrossRef]
- Tian, X.; Yu, H.; Li, D.; Jin, G.; Dai, S.; Gong, P.; Kong, C.; Wang, X. The miR-5694/AF9/Snail Axis Provides Metastatic Advantages and a Therapeutic Target in Basal-like Breast Cancer. Mol. Ther. 2021, 29, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Lu, J.; Zheng, Y.; Wang, P.; Shen, Q. MiR-320a inhibits malignant phenotype of melanoma cells via targeting PBX3. J. BUON 2020, 25, 2071–2077. [Google Scholar]
- Li, Y.; Liu, H.; Shao, J.; Xing, G. miR-320a serves as a negative regulator in the progression of gastric cancer by targeting RAB14. Mol. Med. Rep. 2017, 16, 2652–2658. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yue, L.; Xu, H.; Li, N.; Li, J.; Zhang, Z.; Zhao, R.C. Curcumin suppresses osteogenesis by inducing miR-126a-3p and subsequently suppressing the WNT/LRP6 pathway. Aging 2019, 11, 6983–6998. [Google Scholar] [CrossRef]
- Ding, N.; Sun, X.; Wang, T.; Huang, L.; Wen, J.; Zhou, Y. miR378a3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int. J. Mol. Med. 2018, 42, 381–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.B.; Zhang, X.C.; Chen, P.; Ma, L.M.; Shen, Z.Q. miR378a3p inhibits cellular proliferation and migration in glioblastoma multiforme by targeting tetraspanin 17. Oncol. Rep. 2019, 42, 1957–1971. [Google Scholar] [CrossRef]
- Krist, B.; Podkalicka, P.; Mucha, O.; Mendel, M.; Sepiol, A.; Rusiecka, O.M.; Jozefczuk, E.; Bukowska-Strakova, K.; Grochot-Przeczek, A.; Tomczyk, M.; et al. miR-378a influences vascularization in skeletal muscles. Cardiovasc. Res. 2020, 116, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Khoei, S.G.; Sadeghi, H.; Samadi, P.; Najafi, R.; Saidijam, M. Relationship between Sphk1/S1P and microRNAs in human cancers. Biotechnol. Appl. Biochem. 2020. [Google Scholar] [CrossRef]
- Luca, E.; Turcekova, K.; Hartung, A.; Mathes, S.; Rehrauer, H.; Krützfeldt, J. Genetic deletion of microRNA biogenesis in muscle cells reveals a hierarchical non-clustered network that controls focal adhesion signaling during muscle regeneration. Mol. Metab. 2020, 36, 100967. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, L.; Darrabie, M.D.; Mishra, R.; Jacobs, D.O. Removal of Potential Phosphorylation Sites does not Alter Creatine Transporter Response to PKC or Substrate Availability. Cell. Physiol. Biochem. 2015, 37, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Kori, M.; Aydın, B.; Unal, S.; Arga, K.Y.; Kazan, D. Metabolic Biomarkers and Neurodegeneration: A Pathway Enrichment Analysis of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. OMICS 2016, 20, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Zervou, S.; Whittington, H.J.; Russell, A.J.; Lygate, C.A. Augmentation of Creatine in the Heart. Mini Rev. Med. Chem. 2016, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Brault, J.J.; Abraham, K.A.; Terjung, R.L. Muscle creatine uptake and creatine transporter expression in response to creatine supplementation and depletion. J. Appl. Physiol. 2003, 94, 2173–2180. [Google Scholar] [CrossRef] [Green Version]
- Tarnopolsky, M.; Parise, G.; Fu, M.H.; Brose, A.; Parshad, A.; Speer, O.; Wallimann, T. Acute and moderate-term creatine monohydrate supplementation does not affect creatine transporter mRNA or protein content in either young or elderly humans. Mol. Cell. Biochem. 2003, 244, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Li, J.; Cheng, J. Reconstruction of metabolic pathways by combining probabilistic graphical model-based and knowledge-based methods. BMC Proc. 2014, 8, S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndika, J.D.T.; Martinez-Munoz, C.; Anand, N.; van Dooren, S.J.M.; Kanhai, W.; Smith, D.E.C.; Jakobs, C.; Salomons, G.S. Post-transcriptional regulation of the creatine transporter gene: Functional relevance of alternative splicing. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2070–2079. [Google Scholar] [CrossRef]
- Santacruz, L.; Jacobs, D.O. Structural correlates of the creatine transporter function regulation: The undiscovered country. Amino Acids 2016, 48, 2049–2055. [Google Scholar] [CrossRef]
- Shojaiefard, M.; Christie, D.L.; Lang, F. Stimulation of the creatine transporter SLC6A8 by the protein kinase mTOR. Biochem. Biophys. Res. Commun. 2006, 341, 945–949. [Google Scholar] [CrossRef]
- Shojaiefard, M.; Christie, D.L.; Lang, F. Stimulation of the creatine transporter SLC6A8 by the protein kinases SGK1 and SGK3. Biochem. Biophys. Res. Commun. 2005, 334, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Strutz-Seebohm, N.; Shojaiefard, M.; Christie, D.; Tavare, J.; Seebohm, G.; Lang, F. PIKfyve in the SGK1 Mediated Regulation of the Creatine Transporter SLC6A8. Cell. Physiol. Biochem. 2007, 20, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Lang, M.J.; Weisman, L.S. Phosphatidylinositol 3,5-bisphosphate: Regulation of cellular events in space and time. Biochem. Soc. Trans. 2016, 44, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Berwick, D.C.; Dell, G.C.; Welsh, G.I.; Heesom, K.J.; Hers, I.; Fletcher, L.M.; Cooke, F.T.; Tavare, J.M. Protein kinase B phosphorylation of PIKfyve regulates the trafficking of GLUT4 vesicles. J. Cell Sci. 2004, 117, 5985–5993. [Google Scholar] [CrossRef] [Green Version]
- Kreindler, J.L.; Wiemuth, D.; Lott, J.S.; Ly, K.; Ke, Y.; Teesdale-Spittle, P.; Snyder, P.M.; McDonald, F.J. Interaction of Serum- and Glucocorticoid Regulated Kinase 1 (SGK1) with the WW-Domains of Nedd4-2 Is Required for Epithelial Sodium Channel Regulation. PLoS ONE 2010, 5, e12163. [Google Scholar] [CrossRef]
- Dieter, M.; Palmada, M.; Rajamanickam, J.; Aydin, A.; Busjahn, A.; Boehmer, C.; Luft, F.C.; Lang, F. Regulation of Glucose Transporter SGLT1 by Ubiquitin Ligase Nedd4-2 and Kinases SGK1, SGK3, and PKB. Obes. Res. 2004, 12, 862–870. [Google Scholar] [CrossRef]
- Pakladok, T.; Almilaji, A.; Munoz, C.; Alesutan, I.; Lang, F. PIKfyve Sensitivity of hERG Channels. Cell. Physiol. Biochem. 2013, 31, 785–794. [Google Scholar] [CrossRef]
- Almilaji, A.; Sopjani, M.; Elvira, B.; Borras, J.; Dërmaku-Sopjani, M.; Munoz, C.; Warsi, J.; Lang, U.E.; Lang, F. Upregulation of the Creatine Transporter Slc6A8 by Klotho. Kidney Blood Press. Res. 2014, 39, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Sopjani, M.; Dermaku-Sopjani, M. Klotho-Dependent Cellular Transport Regulation. Vitam. Horm. 2016, 101, 59–84. [Google Scholar] [CrossRef]
- Buchanan, S.; Combet, E.; Stenvinkel, P.; Shiels, P.G. Klotho, Aging, and the Failing Kidney. Front. Endocrinol. 2020, 11. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Liang, L.; Li, J.; Luo, D.; Liu, Q.; Cai, S.; Li, X. Klotho negatively regulated aerobic glycolysis in colorectal cancer via ERK/HIF1alpha axis. Cell Commun. Signal. 2018, 16, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, E.L.; Snow, R.J.; Wright, C.R.; Cho, Y.; Wallace, M.A.; Kralli, A.; Russell, A.P. PGC-1alpha and PGC-1beta increase CrT expression and creatine uptake in myotubes via ERRalpha. Biochim. Biophys. Acta 2014, 1843, 2937–2943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zervou, S.; Yin, X.; Nabeebaccus, A.A.; O’Brien, B.A.; Cross, R.L.; McAndrew, D.J.; Atkinson, R.A.; Eykyn, T.R.; Mayr, M.; Neubauer, S.; et al. Proteomic and metabolomic changes driven by elevating myocardial creatine suggest novel metabolic feedback mechanisms. Amino Acids 2016, 48, 1969–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Hernández, T.; Haucke, V.; Maritzen, T. Endocytosis in the adaptation to cellular stress. Cell Stress 2020, 4, 230–247. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.S.; Tian, X.; Benovic, J.L. Role of beta-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr. Opin. Cell Biol. 2014, 27, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, S.M.; Lee, R.T. Thioredoxin and Thioredoxin Target Proteins: From Molecular Mechanisms to Functional Significance. Antioxid. Redox Signal. 2013, 18, 1165–1207. [Google Scholar] [CrossRef] [Green Version]
- Patwari, P.; Lee, R.T. An expanded family of arrestins regulate metabolism. Trends Endocrinol. Metab. 2012, 23, 216–222. [Google Scholar] [CrossRef] [Green Version]
- Patwari, P.; Chutkow, W.A.; Cummings, K.; Verstraeten, V.L.; Lammerding, J.; Schreiter, E.R.; Lee, R.T. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 2009, 284, 24996–25003. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, A.F.; Schmidt, M.C. AMPK-Mediated Regulation of Alpha-Arrestins and Protein Trafficking. Int. J. Mol. Sci. 2019, 20, 515. [Google Scholar] [CrossRef] [Green Version]
- Spindel, O.N.; World, C.; Berk, B.C. Thioredoxin Interacting Protein: Redox Dependent and Independent Regulatory Mechanisms. Antioxid. Redox Signal. 2012, 16, 587–596. [Google Scholar] [CrossRef]
- Kommaddi, R.P.; Shenoy, S.K. Arrestins and Protein Ubiquitination. Prog. Mol. Biol. Transl. Sci. 2013, 118, 175–204. [Google Scholar] [PubMed]
- Prosser, D.C.; Pannunzio, A.E.; Brodsky, J.L.; Thorner, J.; Wendland, B.; O′Donnell, A.F. α-Arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis. J. Cell Sci. 2015, 128, 4220–4234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallows, K.R.; Bhalla, V.; Oyster, N.M.; Wijngaarden, M.A.; Lee, J.K.; Li, H.; Chandran, S.; Xia, X.; Huang, Z.; Chalkley, R.J.; et al. Phosphopeptide Screen Uncovers Novel Phosphorylation Sites of Nedd4-2 That Potentiate Its Inhibition of the Epithelial Na+ Channel. J. Biol. Chem. 2010, 285, 21671–21678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, H.; Krist, D.T.; Statsyuk, A.V. Crosstalk between kinases and Nedd4 family ubiquitin ligases. Mol. BioSyst. 2014, 10, 1643–1657. [Google Scholar] [CrossRef]
- Shojaiefard, M.; Hosseinzadeh, Z.; Bhavsar, S.K.; Lang, F. Downregulation of the creatine transporter SLC6A8 by JAK2. J. Membr. Biol. 2012, 245, 157–163. [Google Scholar] [CrossRef]
- Fezai, M.; Warsi, J.; Lang, F. Regulation of the Na+,Cl- Coupled Creatine Transporter CreaT (SLC6A8) by the Janus Kinase JAK3. Neurosignals 2015, 23, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, W.; Vinnakota, S.; Qian, X.; Kunze, D.L.; Sarkar, H.K. Molecular Characterization of the Human CRT-1 Creatine Transporter Expressed in Xenopus Oocytes. Arch. Biochem. Biophys. 1999, 361, 75–84. [Google Scholar] [CrossRef]
- Wu, N.; Zheng, B.; Shaywitz, A.; Dagon, Y.; Tower, C.; Bellinger, G.; Shen, C.-H.; Wen, J.; Asara, J.; McGraw, T.E.; et al. AMPK-Dependent Degradation of TXNIP upon Energy Stress Leads to Enhanced Glucose Uptake via GLUT1. Mol. Cell 2013, 49, 1167–1175. [Google Scholar] [CrossRef] [Green Version]
- Darrabie, M.D.; Arciniegas, A.J.; Mishra, R.; Bowles, D.E.; Jacobs, D.O.; Santacruz, L. AMPK and substrate availability regulate creatine transport in cultured cardiomyocytes. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E870–E876. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Thali, R.F.; Smolak, C.; Gong, F.; Alzamora, R.; Wallimann, T.; Scholz, R.; Pastor-Soler, N.M.; Neumann, D.; Hallows, K.R. Regulation of the creatine transporter by AMP-activated protein kinase in kidney epithelial cells. Am. J. Physiol. Ren. Physiol. 2010, 299, F167–F177. [Google Scholar] [CrossRef] [Green Version]
- Galcheva-Gargova, Z.; Derijard, B.; Wu, I.H.; Davis, R.J. An osmosensing signal transduction pathway in mammalian cells. Science 1994, 265, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Fezai, M.; Elvira, B.; Borras, J.; Ben-Attia, M.; Hoseinzadeh, Z.; Lang, F. Negative regulation of the creatine transporter SLC6A8 by SPAK and OSR1. Kidney Blood Press. Res. 2014, 39, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Zhang, J.; Khanna, A.; Hochdorfer, T.; Shang, Y.; Kahle, K.T. The WNK-SPAK/OSR1 pathway: Master regulator of cation-chloride cotransporters. Sci. Signal. 2014, 7, re3. [Google Scholar] [CrossRef] [PubMed]
- Zwiewka, M.; Nodzyński, T.; Robert, S.; Vanneste, S.; Friml, J. Osmotic Stress Modulates the Balance between Exocytosis and Clathrin-Mediated Endocytosis in Arabidopsis thaliana. Mol. Plant 2015, 8, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, S.F.; Kapus, A.; Hoffmann, E.K. Osmosensory Mechanisms in Cellular and Systemic Volume Regulation. J. Am. Soc. Nephrol. 2011, 22, 1587–1597. [Google Scholar] [CrossRef] [Green Version]
- Morbach, S.; Krämer, R. Osmoregulation and osmosensing by uptake carriers for compatible solutes in bacteria. In Molecular Mechanisms Controlling Transmembrane Transport; Springer: Berlin/Heidelberg, Germany, 2004; pp. 121–153. [Google Scholar]
- Wood, J.M. Bacterial Osmosensing Transporters. Methods Enzymol. 2007, 428, 77–107. [Google Scholar]
- El-Kasaby, A.; Kasture, A.; Koban, F.; Hotka, M.; Asjad, H.M.M.; Kubista, H.; Freissmuth, M.; Sucic, S. Rescue by 4-phenylbutyrate of several misfolded creatine transporter-1 variants linked to the creatine transporter deficiency syndrome. Neuropharmacology 2019, 161, 107572. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Groenendyk, J.; Michalak, M. Glycoprotein Quality Control and Endoplasmic Reticulum Stress. Molecules 2015, 20, 13689–13704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farr, C.V.; El-Kasaby, A.; Freissmuth, M.; Sucic, S. The Creatine Transporter Unfolded: A Knotty Premise in the Cerebral Creatine Deficiency Syndrome. Front. Synaptic Neurosci. 2020, 12, 588954. [Google Scholar] [CrossRef]
- Niculescu, A.B. Convergent functional genomics of stem cell-derived cells. Transl. Psychiatry 2013, 3, e305. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dugourd, A.; Kuppe, C.; Sciacovelli, M.; Gjerga, E.; Gabor, A.; Emdal, K.B.; Vieira, V.; Bekker-Jensen, D.B.; Kranz, J.; Bindels, E.M.J.; et al. Causal integration of multi-omics data with prior knowledge to generate mechanistic hypotheses. Mol. Syst. Biol. 2021, 17, e9730. [Google Scholar] [CrossRef] [PubMed]





| Organism | Reference | GEO Number | Design | Creatine | Control | Platform |
|---|---|---|---|---|---|---|
| Human | [70] | GSE7877 | Expression profiling of vastus lateralis muscle in a randomized, placebo- controlled, crossover, double-blind design in young, healthy, non-obese men supplemented with CrM vs. placebo (dextrose) for ten days | 12 | 12 | Buck Institute_Homo sapiens_25K_verC |
| Mouse | [71] | GSE5140 | Analysis of brains of C57Bl/6J animals fed a Cr-supplemented diet for six months | 6 | 7 | Affymetrix Mouse Genome 430 2.0 Array |
| Mouse | [72] | GSE42356 | 3T3 fibroblasts overexpressing CRT were treated with 5mM CrM | 3 | 3 | Illumina MouseWG-6 v2.0 expression beadchip |
| Category | GO ID | Term | Adjusted p-Value |
|---|---|---|---|
| Biological Process | GO:0030879 | Mammary gland development | 0.00538 |
| GO:0043069 | Negative regulation of programmed cell death | 0.01899 | |
| GO:0045765 | Regulation of angiogenesis | 0.03483 | |
| GO:0071542 | Dopaminergic neuron differentiation | 0.05505 | |
| GO:0001934 | Positive regulation of protein phosphorylation | 0.05505 | |
| Cellular Component | GO:0005664 | Nuclear origin of replication recognition complex | 0.2386 |
| GO:0005682 | U5 snRNP | 0.2386 | |
| GO:0031904 | Endosome lumen | 0.2386 | |
| GO:0046540 | U4/U6 x U5 tri-snRNP complex | 0.2386 | |
| GO:0005637 | Nuclear inner membrane | 0.2386 | |
| Molecular Function | GO:0035198 | miRNA binding | 0.05290 |
| GO:0036002 | Pre-mRNA binding | 0.05290 | |
| GO:0048365 | Rac GTPase binding | 0.05290 | |
| GO:0031593 | Polyubiquitin-modification-dependent protein binding | 0.05290 | |
| GO:0001664 | G-protein-coupled receptor binding | 0.05290 | |
| Database | miRBase Accession | Description | Adjusted p-Value |
| miRTarBase | MIMAT0000416 | Mature sequence Homo sapiens miR-1-3p | 0.0942 |
| MIMAT0000275 | Mature sequence Homo sapiens miR-218-5p | 0.1405 | |
| MIMAT0000447 | Mature sequence Homo sapiens miR-134-5p | 0.1513 | |
| MIMAT0022487 | Mature sequence Homo sapiens miR-5694 | 0.1961 | |
| MI0000542 | Stem loop sequence Homo sapiens miR-320a | 0.2492 |
| MicroRNA | Relevant Information |
|---|---|
| miR-1-3p | Suppresses the proliferation of hepatocellular carcinoma [129] and slows the proliferation and invasion of gastric [130] and lung adenocarcinoma [131]. |
| miR-218-5p | Significantly upregulated during myogenic differentiation after activating the IGF-1 and MAPK/ERK pathways [132]. |
| miR-134-5p | Lower levels are found in prostate cancer compared to benign prostatic hyperplasia [133]. In addition, it might have neuroprotective effects by regulating the miR-134-5p/CREB pathway in both humans and mice [134]. |
| miR-5694 | Mediates downregulation of AF9 (a subunit of the super elongation complex and associates with the histone methyltransferases) and provides metastatic advantages in basal-like breast cancer cells [135]. |
| miR-320a | Although associated with certain types of cancer, it has been shown to inhibit the proliferation and progression of melanoma [136] and gastric adenocarcinoma [137]. |
| miR-200b-3p | Higher expression is found in prostate cancer compared to benign prostatic hyperplasia [133]. |
| miR-126a-3p | It targets low-density lipoprotein-receptor-related protein 1 and blocks WNT signaling, which partially explain the anti-tumor effects of curcumin [138]. |
| miR-378a-3p | Exhibits tumor-suppressive and anti-metastatic effects in esophageal squamous cell carcinoma [139] and glioblastoma multiforme [140]; however, miR-378a might also have a pro-angiogenic effect on myoblasts and control vascularization of skeletal muscle [141]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonilla, D.A.; Moreno, Y.; Rawson, E.S.; Forero, D.A.; Stout, J.R.; Kerksick, C.M.; Roberts, M.D.; Kreider, R.B. A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation. Nutrients 2021, 13, 2521. https://doi.org/10.3390/nu13082521
Bonilla DA, Moreno Y, Rawson ES, Forero DA, Stout JR, Kerksick CM, Roberts MD, Kreider RB. A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation. Nutrients. 2021; 13(8):2521. https://doi.org/10.3390/nu13082521
Chicago/Turabian StyleBonilla, Diego A., Yurany Moreno, Eric S. Rawson, Diego A. Forero, Jeffrey R. Stout, Chad M. Kerksick, Michael D. Roberts, and Richard B. Kreider. 2021. "A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation" Nutrients 13, no. 8: 2521. https://doi.org/10.3390/nu13082521
APA StyleBonilla, D. A., Moreno, Y., Rawson, E. S., Forero, D. A., Stout, J. R., Kerksick, C. M., Roberts, M. D., & Kreider, R. B. (2021). A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation. Nutrients, 13(8), 2521. https://doi.org/10.3390/nu13082521