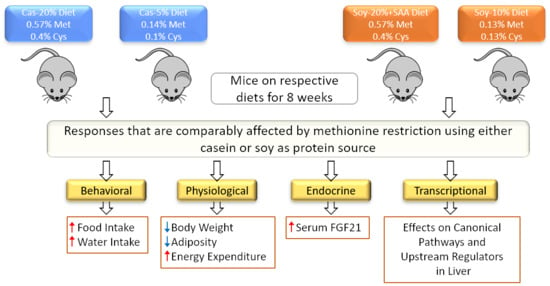
The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Diets and Animals
2.2. Analysis of Energy Expenditure
2.3. Serum Metabolite Analyses
2.4. RNA Isolation and qPCR of MR Target Genes
2.5. RNAseq Analysis
2.6. Bioinformatics Analysis
2.7. Data Analysis
3. Results
3.1. Effects of Protein Restriction on Energy Balance
3.2. Transcriptional Effects of Protein Restriction in Liver and Adipose Tissue
3.3. Differential Gene Expression in the Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anthony, T.G.; Morrison, C.D.; Gettys, T.W. Remodeling of lipid metabolism by dietary restriction of essential amino acids. Diabetes 2013, 62, 2635–2644. [Google Scholar] [CrossRef] [Green Version]
- Forney, L.A.; Stone, K.P.; Wanders, D.; Gettys, T.W. Sensing and signaling mechanisms linking dietary methionine restriction to the behavioral and physiological components of the response. Front. Neuroendocrinol. 2018, 51, 36–45. [Google Scholar] [CrossRef]
- Ables, G.P.; Johnson, J.E. Pleiotropic responses to methionine restriction. Exp. Gerontol. 2017, 94, 83–88. [Google Scholar] [CrossRef]
- Hill, C.M.; Berthoud, H.R.; Munzberg, H.; Morrison, C.D. Homeostatic sensing of dietary protein restriction: A case for FGF21. Front. Neuroendocrinol. 2018, 51, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Yap, Y.W.; Rusu, P.M.; Chan, A.Y.; Fam, B.C.; Jungmann, A.; Solon-Biet, S.M.; Barlow, C.K.; Creek, D.J.; Huang, C.; Schittenhelm, R.B.; et al. Restriction of essential amino acids dictates the systemic metabolic response to dietary protein dilution. Nat. Commun 2020, 11, 2894. [Google Scholar] [CrossRef]
- Otranto, M.; Souza-Netto, I.; Aguila, M.B.; Monte-Alto-Costa, A. Male and female rats with severe protein restriction present delayed wound healing. Appl. Physiol. Nutr. Metab. 2009, 34, 1023–1031. [Google Scholar] [CrossRef]
- White, B.D.; Porter, M.H.; Martin, R.J. Protein selection, food intake, and body composition in response to the amount of dietary protein. Physiol. Behav. 2000, 69, 383–389. [Google Scholar] [CrossRef]
- Forney, L.A.; Wanders, D.; Stone, K.P.; Pierse, A.; Gettys, T.W. Concentration-dependent linkage of dietary methionine restriction to the components of its metabolic phenotype. Obesity 2017, 25, 730–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothwell, N.J.; Stock, M.J.; Tyzbir, R.S. Mechanisms of thermogenesis induced by low protein diets. Metabolism 1983, 32, 257–261. [Google Scholar] [CrossRef]
- Rothwell, N.J.; Stock, M.J. Influence of carbohydrate and fat intake on diet-induced thermogenesis and brown fat activity in rats fed low protein diets. J. Nutr. 1987, 117, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Laeger, T.; Henagan, T.M.; Albarado, D.C.; Redman, L.M.; Bray, G.A.; Noland, R.C.; Munzberg, H.; Hutson, S.M.; Gettys, T.W.; Schwartz, M.W.; et al. FGF21 is an endocrine signal of protein restriction. J. Clin. Investig. 2014, 124, 3913–3922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laeger, T.; Albarado, D.C.; Burke, S.J.; Trosclair, L.; Hedgepeth, J.W.; Berthoud, H.R.; Gettys, T.W.; Collier, J.J.; Munzberg, H.; Morrison, C.D. Metabolic Responses to Dietary Protein Restriction Require an Increase in FGF21 that Is Delayed by the Absence of GCN2. Cell Rep. 2016, 16, 707–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maida, A.; Zota, A.; Sjoberg, K.A.; Schumacher, J.; Sijmonsma, T.P.; Pfenninger, A.; Christensen, M.M.; Gantert, T.; Fuhrmeister, J.; Rothermel, U.; et al. A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J. Clin. Investig. 2016, 126, 3263–3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, E.K.; Banks, R.; Cook, C.; Hill, S.; Morrice, N.; Grant, L.; Mody, N.; Delibegovic, M. Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci. Rep. 2017, 7, 9977. [Google Scholar] [CrossRef]
- Du, Y.; Meng, Q.; Zhang, Q.; Guo, F. Isoleucine or valine deprivation stimulates fat loss via increasing energy expenditure and regulating lipid metabolism in WAT. Amino Acids 2012, 43, 725–734. [Google Scholar] [CrossRef]
- Wanders, D.; Stone, K.P.; Dille, K.; Simon, J.; Pierse, A.; Gettys, T.W. Metabolic responses to dietary leucine restriction involve remodeling of adipose tissue and enhanced hepatic insulin signaling. Biofactors 2015, 41, 391–402. [Google Scholar] [CrossRef] [Green Version]
- Hasek, B.E.; Boudreau, A.; Shin, J.; Feng, D.; Hulver, M.; Van, N.T.; Laque, A.; Stewart, L.K.; Stone, K.P.; Wanders, D.; et al. Remodeling the integration of lipid metabolism between liver and adipose tissue by dietary methionine restriction in rats. Diabetes 2013, 62, 3362–3372. [Google Scholar] [CrossRef] [Green Version]
- Wanders, D.; Burk, D.H.; Cortez, C.C.; Van, N.T.; Stone, K.P.; Baker, M.; Mendoza, T.; Mynatt, R.L.; Gettys, T.W. UCP1 is an essential mediator of the effects of methionine restriction on energy balance but not insulin sensitivity. FASEB J. 2015, 29, 2603–2615. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Stone, K.P.; Forney, L.A.; Sims, L.C.; Gutierrez, G.C.; Ghosh, S.; Gettys, T.W. Implementation of dietary methionine restriction using casein after selective, oxidative deletion of methionine. iScience 2021, 24, 102470. [Google Scholar] [CrossRef]
- Stone, K.P.; Ghosh, S.; Kovalik, J.P.; Orgeron, M.; Wanders, D.; Sims, L.C.; Gettys, T.W. The acute transcriptional responses to dietary methionine restriction are triggered by inhibition of ternary complex formation and linked to Erk1/2, mTOR, and ATF4. Sci. Rep. 2021, 11, 3765. [Google Scholar] [CrossRef]
- Forney, L.A.; Stone, K.P.; Gibson, A.N.; Vick, A.M.; Sims, L.C.; Fang, H.; Gettys, T.W. Sexually Dimorphic Effects of Dietary Methionine Restriction are Dependent on Age when the Diet is Introduced. Obesity 2020, 28, 581–589. [Google Scholar] [CrossRef]
- Forney, L.A.; Fang, H.; Sims, L.C.; Stone, K.P.; Vincik, L.Y.; Vick, A.M.; Gibson, A.N.; Burk, D.H.; Gettys, T.W. Dietary Methionine Restriction Signals to the Brain Through Fibroblast Growth Factor 21 to Regulate Energy Balance and Remodeling of Adipose Tissue. Obesity 2020, 28, 1912–1921. [Google Scholar] [CrossRef]
- Fang, H.; Stone, K.P.; Ghosh, S.; Forney, L.A.; Sims, L.C.; Vincik, L.; Gettys, T.W. Hepatic Nfe2l2 Is Not an Essential Mediator of the Metabolic Phenotype Produced by Dietary Methionine Restriction. Nutrients 2021, 13, 1788. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, A.; Yekutieli, D.; Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef]
- Wanders, D.; Forney, L.A.; Stone, K.P.; Hasek, B.E.; Johnson, W.D.; Gettys, T.W. The Components of Age-Dependent Effects of Dietary Methionine Restriction on Energy Balance in Rats. Obesity 2018, 26, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.M.; Laeger, T.; Dehner, M.; Albarado, D.C.; Clarke, B.; Wanders, D.; Burke, S.J.; Collier, J.J.; Qualls-Creekmore, E.; Solon-Biet, S.M.; et al. FGF21 Signals Protein Status to the Brain and Adaptively Regulates Food Choice and Metabolism. Cell Rep. 2019, 27, 2934–2947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malloy, V.L.; Krajcik, R.A.; Bailey, S.J.; Hristopoulos, G.; Plummer, J.D.; Orentreich, N. Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell 2006, 5, 305–314. [Google Scholar] [CrossRef]
- Orgeron, M.; Stone, K.P.; Wanders, D.; Cortez, C.C.; Van, N.T.; Gettys, T.W. The Impact of Dietary Methionine Restriction on Biomarkers of Metabolic Health. In Progress in Molecular Biology and Translational Science; Tao, Y., Ed.; Academic Press: Auburn, AL, USA, 2014; Volume 121, pp. 351–376. [Google Scholar]
- Soultoukis, G.A.; Partridge, L. Dietary Protein, Metabolism, and Aging. Annu. Rev. Biochem. 2016, 85, 5–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzaei, H.; Raynes, R.; Longo, V.D. The conserved role of protein restriction in aging and disease. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 74–79. [Google Scholar] [CrossRef] [Green Version]
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Solon-Biet, S.M.; McMahon, A.C.; Ballard, J.W.; Ruohonen, K.; Wu, L.E.; Cogger, V.C.; Warren, A.; Huang, X.; Pichaud, N.; Melvin, R.G.; et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014, 19, 418–430. [Google Scholar] [CrossRef] [Green Version]
- Hasek, B.E.; Stewart, L.K.; Henagan, T.M.; Boudreau, A.; Lenard, N.R.; Black, C.; Shin, J.; Huypens, P.; Malloy, V.L.; Plaisance, E.P.; et al. Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R728–R739. [Google Scholar] [CrossRef] [Green Version]
- Richie, J.P., Jr.; Leutzinger, Y.; Parthasarathy, S.; Malloy, V.; Orentreich, N.; Zimmerman, J.A. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994, 8, 1302–1307. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, J.D.; Martin, J.J.; Harris, B.J. Methionine metabolism in mammals. The methionine-sparing effect of cystine. J. Biol. Chem. 1988, 263, 11750–11754. [Google Scholar] [CrossRef]
- Aoyama, S.; Hirooka, R.; Shimoda, T.; Shibata, S. Effect of different sources of dietary protein on muscle hypertrophy in functionally overloaded mice. Biochem. Biophys. Rep. 2019, 20, 100686. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Wang, D.; Wang, H.; Chen, T.; Luo, J.; Xi, Q.; Sun, J.; Zhang, Y. Dietary Soy Protein Isolate Attenuates Intestinal Immunoglobulin and Mucin Expression in Young Mice Compared with Casein. Nutrients 2020, 12, 2739. [Google Scholar] [CrossRef]
- Blais, A.; Chaumontet, C.; Azzout-Marniche, D.; Piedcoq, J.; Fromentin, G.; Gaudichon, C.; Tome, D.; Even, P.C. Low-protein diet-induced hyperphagia and adiposity are modulated through interactions involving thermoregulation, motor activity, and protein quality in mice. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E139–E151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaumontet, C.; Azzout-Marniche, D.; Blais, A.; Piedcoq, J.; Tome, D.; Gaudichon, C.; Even, P.C. Low-protein and methionine, high-starch diets increase energy intake and expenditure, increase FGF21, decrease IGF-1, and have little effect on adiposity in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 316, R486–R501. [Google Scholar] [CrossRef]
- Ghosh, S.; Forney, L.A.; Wanders, D.; Stone, K.P.; Gettys, T.W. An integrative analysis of tissue-specific transcriptomic and metabolomic responses to short-term dietary methionine restriction in mice. PLoS ONE 2017, 12, e0177513. [Google Scholar] [CrossRef]
- Ghosh, S.; Wanders, D.; Stone, K.P.; Van, N.T.; Cortez, C.C.; Gettys, T.W. A systems biology analysis of the unique and overlapping transcriptional responses to caloric restriction and dietary methionine restriction in rats. FASEB J. 2014, 28, 2577–2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrone, C.E.; Mattocks, D.A.; Plummer, J.D.; Chittur, S.V.; Mohney, R.; Vignola, K.; Orentreich, D.S.; Orentreich, N. Genomic and metabolic responses to methionine-restricted and methionine-restricted, cysteine-supplemented diets in Fischer 344 rat inguinal adipose tissue, liver and quadriceps muscle. Lifestyle Genom. 2012, 5, 132–157. [Google Scholar] [CrossRef]
- Ascencio, C.; Torres, N.; Isoard-Acosta, F.; Gomez-Perez, F.J.; Hernandez-Pando, R.; Tovar, A.R. Soy protein affects serum insulin and hepatic SREBP-1 mRNA and reduces fatty liver in rats. J. Nutr. 2004, 134, 522–529. [Google Scholar] [CrossRef]
- Lavigne, C.; Marette, A.; Jacques, H. Cod and soy proteins compared with casein improve glucose tolerance and insulin sensitivity in rats. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E491–E500. [Google Scholar] [CrossRef]
- Iwasaki, K.; Gleiser, C.A.; Masoro, E.J.; McMahan, C.A.; Seo, E.J.; Yu, B.P. The influence of dietary protein source on longevity and age-related disease processes of Fischer rats. J. Gerontol. 1988, 43, B5–B12. [Google Scholar] [CrossRef]
- Berry, S.A.; Brown, C.S.; Greene, C.; Camp, K.M.; McDonough, S.; Bocchini, J.A. Medical Foods for Inborn Errors of Metabolism: History, Current Status, and Critical Need. Pediatrics 2020, 145, e20192261. [Google Scholar] [CrossRef]
- Plaisance, E.P.; Greenway, F.L.; Boudreau, A.; Hill, K.L.; Johnson, W.D.; Krajcik, R.A.; Perrone, C.E.; Orentreich, N.; Cefalu, W.T.; Gettys, T.W. Dietary methionine restriction increases fat oxidation in obese adults with metabolic syndrome. J. Clin. Endocrinol. Metab. 2011, 96, E836–E840. [Google Scholar] [CrossRef]
- Elshorbagy, A.K.; Valdivia-Garcia, M.; Mattocks, D.A.; Plummer, J.D.; Smith, A.D.; Drevon, C.A.; Refsum, H.; Perrone, C.E. Cysteine supplementation reverses methionine restriction effects on rat adiposity: Significance of stearoyl-coenzyme A desaturase. J. Lipid Res. 2011, 52, 104–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshorbagy, A.K.; Valdivia-Garcia, M.; Mattocks, D.A.; Plummer, J.D.; Orentreich, D.S.; Orentreich, N.; Refsum, H.; Perrone, C.E. Effect of taurine and N-acetylcysteine on methionine restriction-mediated adiposity resistance. Metabolism 2013, 62, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Stone, K.P.; Forney, L.A.; Cortez, C.C.; Dille, K.N.; Simon, J.; Xu, M.; Hotard, E.C.; Nikonorova, I.A.; Pettit, A.P.; et al. Role of GCN2-independent signaling through a non-canonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 2016, 65, 1499–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Cas-20% | Cas-5% | Soy-20% 1 | Soy-20% + SAA | Soy-10% | |
|---|---|---|---|---|---|
| Casein or soy protein, g | 200 | 50 | 200 | 200 | 115 |
| Grams of added Met | 0 | 0 | 0 | 3.53 2 | 0 |
| Final [Met], gm per 100 g diet | 0.57 | 0.14 | 0.22 | 0.57 | 0.13 |
| Grams of added Cys | 3.00 3 | 0.75 4 | 0 | 1.91 5 | 0 |
| Final [Cys], gm per 100 g diet | 0.41 | 0.10 | 0.22 | 0.40 | 0.13 |
| Corn Starch, g | 376 | 485 | 376 | 370 | 426 |
| Maltodextrin 10, g | 125 | 150 | 125 | 125 | 150 |
| Sucrose, g | 107 | 107 | 107 | 107 | 107 |
| Cellulose, g | 50 | 50 | 50 | 50 | 50 |
| Soybean Oil, g | 25 | 25 | 25 | 25 | 25 |
| Lard, g | 75 | 75 | 69.6 | 69.6 | 72.3 |
| Mineral Mix S10022C, g | 3.5 | 3.5 | 3.5 | 3.5 | 3.5 |
| Calcium Carbonate, g | 12.5 | 8.7 | 10.6 | 10.6 | 9.3 |
| Calcium Phosphate, g | 0 | 5.3 | 0 | 0 | 3.0 |
| Potassium Citrate, 1 H2O, g | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Potassium Phosphate, g | 6.9 | 6.9 | 6.9 | 6.9 | 6.9 |
| Sodium Chloride, g | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 |
| Vitamin Mix V10037, g | 10 | 10 | 10 | 10 | 10 |
| Choline Bitartrate, g | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Total | 1001 | 985 | 991 | 991 | 986 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Stone, K.P.; Ghosh, S.; Forney, L.A.; Gettys, T.W. The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein. Nutrients 2021, 13, 2609. https://doi.org/10.3390/nu13082609
Fang H, Stone KP, Ghosh S, Forney LA, Gettys TW. The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein. Nutrients. 2021; 13(8):2609. https://doi.org/10.3390/nu13082609
Chicago/Turabian StyleFang, Han, Kirsten P. Stone, Sujoy Ghosh, Laura A. Forney, and Thomas W. Gettys. 2021. "The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein" Nutrients 13, no. 8: 2609. https://doi.org/10.3390/nu13082609
APA StyleFang, H., Stone, K. P., Ghosh, S., Forney, L. A., & Gettys, T. W. (2021). The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein. Nutrients, 13(8), 2609. https://doi.org/10.3390/nu13082609