Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Dry-Cured Pork Loin during Aging and Gastrointestinal Digestion
Abstract
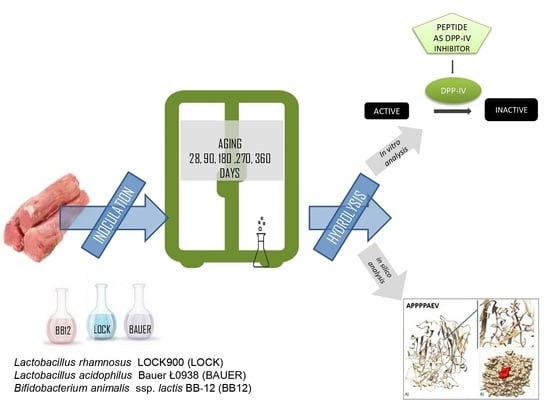
:1. Introduction
2. Materials and Methods
2.1. Dry-Cured Loins Preparation
2.2. Muscle Protein Extraction and Gastro-Intestinal Digestion
2.3. Assessment of Peptides Content during the Digestion
2.4. Dipeptidyl-Peptidase IV Inhibitor Screening Activity Determination
2.5. Spectrometric Peptides (>7 kDa) Identification and Computational Study
2.6. In Silico Prediction for Activity of the Identified Peptides
2.7. Docking Study
2.8. Statistical Analysis
3. Results
3.1. Evaluation of the DPP-IVi of the Intact Proteins (Extracts) from Dry-Cured Pork Loins
3.2. Evaluation of the DPP-IV Inhibiting Activity of the Extracts during Simulated Gastro-Intestinal Digestion (SGID)
3.3. Peptide Profile of the Hydrolysates by LC–MS/MS and Computational Study
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peighambardoust, S.H.; Karami, Z.; Pateiro, M.; Lorenzo, J.M. A Review on Health-Promoting, Biological, and Functional Aspects of Bioactive Peptides in Food Applications. Biomolecules 2021, 11, 631. [Google Scholar] [CrossRef]
- Chai, K.F.; Voo, A.Y.H.; Chen, W.N. Bioactive peptides from food fermentation: A comprehensive review of their sources, bioactivities, applications, and future development. Comp. Rev. Food Sci. Food Saf. 2020, 19, 3825–3885. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Gallego, M.; Reig, M.; Aristoy, M.C.; Mora, L. Bioactive peptides generated in the processing of dry-cured ham. Food Chem. 2020, 321, 126689. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tomé, S.; Hernández-Ledesma, B. Gastrointestinal digestion of food proteins under the effects of released bioactive peptides on digestive health. Mol. Nutr. Food Res. 2020, 64, 2000401. [Google Scholar] [CrossRef]
- Toldra, F.; Gallego, M.; Reig, M.; Aristoy, M.C.; Mora, L. Recent progress in enzymatic release of peptides in foods of animal origin and assessment of bioactivity. J. Agric. Food Chem. 2020, 68, 12842–12855. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; Hayes, M. Bioactive peptides from meat muscle and by-products: Generation, functionality and application as functional ingredients. Meat Sci. 2014, 98, 227–239. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Fujita, H.; Matoba, N.; Takenaka, Y.; Yamamoto, T.; Yamauchi, R.; Tsuruki, H.; Takahata, K. Bioactive peptides derived from food proteins preventing lifestyle-related diseases. Biofactors 2000, 12, 143–146. [Google Scholar] [CrossRef]
- Cicero, A.F.; Fogacci, F.; Colletti, A. Potential role of bioactive peptides in prevention and treatment of chronic diseases: A narrative review. Br. J. Pharmacol. 2017, 174, 1378–1394. [Google Scholar] [CrossRef]
- Chaudhury, A.; Duvoor, C.; Reddy Dendi, V.S.; Kraleti, S.; Chada, A.; Ravilla, R.; Marco, A.; Shekhawat, N.S.; Montales, M.T.; Kuriakose, K.; et al. Clinical review of antidiabetic drugs: Implications for type 2 diabetes mellitus management. Front. Endocrinol. 2017, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [Green Version]
- Molina, I.; Toldrá, F. Detection of proteolytic activity in microorganisms isolated from dry-cured ham. J. Food Sci. 1992, 57, 1308–1310. [Google Scholar] [CrossRef]
- Fadda, S.; Sanz, Y.; Vignolo, G.; Aristoy, M.C.; Oliver, G.; Toldrá, F. Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 1999, 65, 3540–3546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kęska, P.; Stadnik, J. Stability of antiradical activity of protein extracts and hydrolysates from dry-cured pork loins with probiotic strains of LAB. Nutrients 2018, 10, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid sensitive method for the quantification of microgram quantities of protein utilising the principle of protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Escudero, E.; Santandreu, M.A.; Toldra, F. Characterization of peptides released by in vitro digestion of pork meat. J. Agric. Food Chem. 2010, 58, 5160–5165. [Google Scholar] [CrossRef] [PubMed]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- BIOPEP-UWM. Available online: https://biochemia.uwm.edu.pl/biopep-uwm/ (accessed on 1 November 2021).
- ToxinPred. Available online: http://crdd.osdd.net/raghava/toxinpred/ (accessed on 1 November 2021).
- Aller Top. Available online: https://www.ddg-pharmfac.net/AllerTOP/ (accessed on 1 November 2021).
- CPPpred. Available online: http://distilldeep.ucd.ie/CPPpred/ (accessed on 1 November 2021).
- iDPPIV-SCM. Available online: http://camt.pythonanywhere.com/iDPPIV-SCM (accessed on 1 November 2021).
- Singh, H.; Singh, S.; Raghava, G.P.S. Peptide secondary structure prediction using evolutionary information. BioRxiv 2019, 558791. [Google Scholar]
- Kurcinski, M.; Ciemny, M.P.; Oleniecki, T.; Kuriata, A.; Badaczewska-Dawid, A.E.; Kolinski, A.; Kmiecik, S. CABS-dock standalone: A toolbox for flexible protein–peptide docking. Bioinformatics 2019, 35, 4170–4172. [Google Scholar] [CrossRef]
- CABSdock. Available online: https://bitbucket.org/lcbio/cabsdock/wiki/Home (accessed on 1 November 2021).
- Kęska, P.; Stadnik, J. Porcine myofibrillar proteins as potential precursors of bioactive peptides–An in silico study. Food Funct. 2016, 7, 2878–2885. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J.; Wójciak, K.M.; Neffe-Skocińska, K. Physico-chemical and proteolytic changes during cold storage of dry-cured pork loins with probiotic strains of LAB. Int. J. Food Sci. Technol. 2020, 55, 1069–1079. [Google Scholar] [CrossRef]
- Kęska, P.; Stadnik, J. Ageing-time dependent changes of angiotensin I-converting enzyme-inhibiting activity of protein hydrolysates obtained from dry-cured pork loins inoculated with probiotic lactic acid bacteria. Int. J. Pept. Res. Ther. 2019, 25, 1173–1185. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, A.; Mudgil, P.; Palakkott, A.; Iratni, R.; Gan, C.Y.; Maqsood, S.; Ayoub, M.A. Molecular basis of the anti-diabetic properties of camel milk through profiling of its bioactive peptides on dipeptidyl peptidase IV (DPP-IV) and insulin receptor activity. J. Dairy Sci. 2021, 104, 61–77. [Google Scholar] [CrossRef]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Production and identification of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from discarded Sardine pilchardus protein. Food Chem. 2020, 328, 127096. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, P.; Kilari, B.P.; Kamal, H.; Olalere, O.A.; FitzGerald, R.J.; Gan, C.Y.; Maqsood, S. Multifunctional bioactive peptides derived from quinoa protein hydrolysates: Inhibition of α-glucosidase, dipeptidyl peptidase-IV and angiotensin I converting enzymes. J. Cereal Sci. 2020, 96, 103130. [Google Scholar] [CrossRef]
- Lacroix, I.M.; Li-Chan, E.C. Evaluation of the potential of dietary proteins as precursors of dipeptidyl peptidase (DPP)-IV inhibitors by an in silico approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef]
- Singla, R.K.; Kumar, R.; Khan, S.; Kumari, K.; Garg, A. Natural Products: Potential Source of DPP-IV Inhibitors. Curr. Protein Pept. Sci. 2019, 20, 1218–1225. [Google Scholar] [CrossRef]
- Martini, S.; Conte, A.; Tagliazucchi, D. Comparative peptidomic profile and bioactivities of cooked beef, pork, chicken and turkey meat after in vitro gastro-intestinal digestion. J. Proteom. 2019, 208, 103500. [Google Scholar] [CrossRef] [PubMed]
- Marušić, N.; Aristoy, M.C.; Toldrá, F. Nutritional pork meat compounds as affected by ham dry-curing. Meat Sci. 2014, 93, 53–60. [Google Scholar] [CrossRef]
- Gallego, M.; Aristoy, M.C.; Toldrá, F. Dipeptidyl peptidase IV inhibitory peptides generated in Spanish dry-cured ham. Meat Sci. 2014, 96, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Dipeptidyl peptidase IV inhibitory properties of a whey protein hydrolysate: Influence of fractionation, stability to simulated gastrointestinal digestion and food–drug interaction. Int. Dairy J. 2013, 32, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.L.; Jao, C.L.; Ho, K.P.; Hsu, K.C. Dipeptidyl-peptidase IV inhibitory activity of peptides derived from tuna cooking juice hydrolysates. Peptides 2012, 35, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; O’Keeffe, M.B.; FitzGerald, R.J. Purification and identification of dipeptidyl peptidase (DPP) IV inhibitory peptides from the macroalga Palmaria palmata. Food Chem. 2015, 172, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Vilcacundo, R.; Martínez-Villaluenga, C.; Hernández-Ledesma, B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J. Funct. Foods 2017, 35, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Goodman, R.E. Biosafety: Evaluation and regulation ofgenetically modified (GM) crops in the United States. J. Huazhong Agric. Univ. 2014, 33, 85–114. [Google Scholar]
- Hayes, M.; Rougé, P.; Barre, A.; Herouet-Guicheney, C.; Roggen, E.L. In silico tools for exploring potential human allergy to proteins. Drug Discov. Today Dis. Models 2015, 17–18, 3–11. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Kim, G.C.; Cheon, D.H.; Lee, Y. Challenge to overcome current limitations of cell-penetrating peptides. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2021, 1869, 140604. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open Source Drug Discovery Consortium; Raghava, G.P. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, I.M.; Li-Chan, E.C. Dipeptidyl peptidase-IV inhibitory activity of dairy protein hydrolysates. Int. Dairy J. 2012, 25, 97–102. [Google Scholar] [CrossRef]
- Power, O.; Nongonierma, A.B.; Jakeman, P.; FitzGerald, R.J. Food protein hydrolysates as a source of dipeptidyl peptidase IV inhibitory peptides for the management of type 2 diabetes. Proc. Nutr. Soc. 2014, 73, 34–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boots, J.-W.P. Protein hydrolysate enriched in peptides inhibiting DPP-IV and their use. U.S. Patent 8273710, 25 September 2012. [Google Scholar]
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.U.; Brandstetter, H. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of bioactive peptides by Lactobacillus species: From gene to application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.P.; Ardö, Y. Variation in aminopeptidase and aminotransferase activities of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 2010, 20, 149–155. [Google Scholar] [CrossRef]




| Factor | Peptides Concentration (mg/mL) | DPP-IVi (%) | ||
|---|---|---|---|---|
| WSF | SSW | WSF | SSW | |
| Intact proteins | ||||
| Treatment (T) | N.S. | *** | *** | *** |
| Storage time (S) | *** | *** | *** | *** |
| TxS | *** | *** | *** | *** |
| Post-gastric | ||||
| Treatment (T) | * | N.S. | *** | *** |
| Storage time (S) | *** | *** | *** | *** |
| TxS | * | ** | *** | *** |
| Post- gastrointestinal | ||||
| Treatment (T) | *** | N.S. | *** | *** |
| Storage time (S) | *** | *** | *** | *** |
| TxS | ** | * | *** | *** |
| After simulated adsorption | ||||
| Treatment (T) | N.S. | N.S. | *** | *** |
| Storage time (S) | *** | * | *** | *** |
| TxS | N.S. | N.S. | *** | *** |
| Time (Day) | Variants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | LOCK | BAUER | BB12 | ||||||
| PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi (%) | PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | ||
| WSF | 28 | 1.46 Ab ± 0.01 | 73.70 Ab ± 0.21 | 1.59 Ab ± 0.11 | 65.10 Cb ± 0.21 | 1.56 Ac ± 0.15 | 70.85 Bc ± 0.208 | 1.48 Ac ± 0.05 | 61.01 Db ± 1.63 |
| 90 | 2.08 Ba ± 0.06 | 85.90 Ba ± 0.14 | 2.18 ABa ± 0.05 | 96.88 Aa ± 0.10 | 2.20 Aba ± 0.01 | 85.90 Ba ± 0.14 | 2.41 Ab ± 0.09 | 85.95 Ba ± 0.14 | |
| 180 | 1.69 Ab ± 0.13 | 84.51 Aa ± 4.31 | 1.35 ABc ± 0.15 | 68.89 Cb ±4.31 | 0.82 Cd ± 0.08 | 78.50 Bab ± 2.98 | 1.14 BCd ± 0.07 | 81.49 ABa ± 1.25 | |
| 270 | 1.42 Db ± 0.02 | 24.56 Bd ± 1.03 | 1.67 Cb ± 0.02 | 23.94 Bc ± 1.03 | 1.86 Bb ± 0.04 | 31.49 Ad ± 1.49 | 2.07 Aa ± 0.06 | 19.63 Cd ±0.80 | |
| 360 | 2.15 Aa ± 0.18 | 36.66 Ac ±2.22 | 2.14 Aa ± 0.03 | 14.99 Cd ± 3.66 | 2.04 Aab ± 0.14 | 27.73 Be ± 2.97 | 1.87 Aa ± 0.11 | 31.49 Cc ± 2.7 | |
| SSF | 28 | 0.42 Bd ± 0.01 | 68.98 Cb ± 0.40 | 0.65 Ac ± 0.03 | 84.97 Aa ± 0.88 | 0.60 Ac ± 0.02 | 73.11 Bb ± 0.98 | 0.41 Bc ± 0.05 | 73.09 Bb ± 0.18 |
| 90 | 1.21 Ba ± 0.04 | 54.80 Bc ± 0.33 | 0.93 Cb ± 0.03 | 51.38 Bb ± 3.78 | 1.15 Ba ± 0.04 | 64.21 Ac ± 3.62 | 1.34 Aa ± 0.03 | 61.32 Ac ± 0.22 | |
| 180 | 1.22 Aa ± 0.05 | 85.37 Aa ± 1.28 | 1.20 Aa ± 0.13 | 85.00 Aba ± 2.65 | 0.81 Bb ± 0.09 | 81.39 Ca ± 0.98 | 1.15 ABb ± 0.03 | 82.46 BCa ± 1.34 | |
| 270 | 0.56 Bc ± 0.001 | 15.73 Cd ± 3.61 | 0.45 Bd ± 0.06 | 30.63 Bc ± 2.20 | 0.79 Ab ± 0.02 | 34.04 ABd ± 2.44 | 0.52 Bd ± 0.03 | 36.01 Ad ± 0.21 | |
| 360 | 0.98 Ab ± 0.03 | 53.06 Ac ± 0.58 | 0.88 Ab ± 0.04 | 11.32 Cd ± 0.06 | 0.50 Bc ± 0.04 | 17.92 Be ± 2.59 | 0.56 Bd ± 0.01 | 10.71 Ce ± 0.52 | |
| Time (Day) | Dipeptidyl-Peptidase-IV-Inhibiting Activity(%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | LOCK | BAUER | BB12 | ||||||
| PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | ||
| Post-gastric | 28 | 1.62 Ab ± 0.22 | 71.66 Ca ± 0.67 | 1.44 Ac ± 0.03 | 69.80 Cb ± 2.42 | 1.45 Ac ± 0.01 | 77.69 Bb ± 0.74 | 1.51 Ad ± 0.05 | 84.01 Ab ± 1.05 |
| 90 | 1.51 Cc ± 0.02 | 48.23 Bd ± 3.12 | 1.54 Cbc ± 0.03 | 42.00 Bd ± 0.25 | 1.92 Ab ± 0.09 | 52.87 Ad ± 1.58 | 2.13 Abc ± 0.07 | 33.36 Ce ± 0.78 | |
| 180 | 2.05 Ab ± 0.14 | 84.51 Db ± 1.54 | 2.45 Aa ± 0.46 | 93.17 Ca ± 0.51 | 2.22 Ab ± 0.23 | 98.7 Aa ± 1.18 | 2.32 Aab ± 0.26 | 95.29 Ba ± 0.95 | |
| 270 | 1.94 Bb ± 0.04 | 64.370 ABc ± 0.834 | 2.00 Bab ± 0.03 | 60.33 Bc ±0.67 | 2.29 Aab ± 0.04 | 60.56 Bc ± 1.68 | 1.99 Bc ± 0.02 | 67.06 Ac ± 1.61 | |
| 360 | 2.68 Aa ± 0.13 | 65.14 Ac ± 6.78 | 2.14 Aa ± 0.03 | 40.93 Cd ± 2.52 | 2.70 Aa ± 0.21 | 51.69 Bd ± 1.76 | 2.52 Aa ± 0.08 | 39.87 Cd ± 0.49 | |
| Post-gastrointestinal | 28 | 1.73 Ac ± 0.003 | 21.99 Bd ± 0.07 | 1.66 Ad ± 0.07 | 20.45 Bd ± 2.05 | 1.71 Ab ± 0.005 | 23.74 Bd ± 0.37 | 1.64 Ac ± 0.10 | 32.00 Ac ± 2.83 |
| 90 | 1.80 Bc ± 0.01 | 53.93 Ab ± 0.81 | 2.00 Ac ± 0.02 | 28.50 Cd ± 5.80 | 1.84 ABb ± 0.06 | 35.04 CDc ± 4.20 | 1.90 ABc ± 0.05 | 39.82 Dc ± 0.77 | |
| 180 | 2.13 Ca ± 0.01 | 86.28 Ba ± 1.01 | 3.11 Ab ± 0.01 | 85.81 Ba ± 0.36 | 2.88 Ba ± 0.06 | 95.94 Aa ± 4.05 | 2.7 Bb ± 0.03 | 94.572 Aa ± 1.67 | |
| 270 | 2.11 Aa ± 0.23 | 82.875 Ba ± 0.36 | 1.94 Ac ± 0.24 | 78.49 Cb ± 1.216 | 2.69 Aa ± 0.26 | 75.982 Bb ±1.87 | 2.21 Ab ± 0.28 | 84.42 Ab ± 2.72 | |
| 360 | 2.31 Ca ± 0.004 | 44.88 Bc ± 1.96 | 3.44 Aa ± 0.02 | 53.63 Ac ± 0.99 | 3.03 Ba ± 0.04 | 31.09 Ccd ± 2.43 | 3.61 Aa ± 0.45 | 44.41 Bc ± 4.17 | |
| After simulated adsorption | 28 | 0.63 ABc ± 0.07 | 35.00 Bb ± 2.83 | 0.54 Bc ± 0.03 | 37.438 Bb ± 3.75 | 0.60 B ± 0.01 | 40.21 Ab ± 0.56 | 0.76 Ac ±0.02 | 37.91 Bb ± 2.49 |
| 90 | 1.27 Ca ± 0.02 | 33.92 Bb ± 1.67 | 1.60 Ba ± 0.01 | 42.32 Ab ± 5.43 | 1.78 A ± 0.01 | 38.28 Ab ± 2.96 | 1.72 Aa ± 0.02 | 42.33 Ab ± 0.14 | |
| 180 | 0.98 Bb ± 0.09 | 73.37 Ba ± 0.95 | 1.44 Aa ± 0.07 | 55.19 Da ± 1.09 | 1.40 A ± 0.03 | 69.17 Ca ± 1.16 | 1.59 Aab ± 0.07 | 81.05 Aa ± 1.48 | |
| 270 | 1.27 Aa ± 0.02 | 25.76 Bb ± 4.79 | 1.25 Aab ± 0.08 | 41.53 Ab ± 0.10 | 1.39 A ± 0.10 | 42.40 Ab ± 1.49 | 1.32 Ab ± 0.05 | 39.80 Ab ± 0.50 | |
| 360 | 0.82 Ac ± 0.04 | 30.44 Ab ± 2.78 | 1.02 Ab ± 0.11 | 20.74 Bc ± 3.21 | 0.64 A ± 0.24 | 11.35 Cc ± 0.158 | 0.75 Ac ± 0.12 | 20.41 Bc ± 2.45 | |
| Time (Day) | Dipeptidyl-Peptidase-IV-Inhibiting Activity (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | LOCK | BAUER | BB12 | ||||||
| PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | PC (mg/mL) | DPP-IVi(%) | ||
| Post-gastric | 28 | 1.31 Ab ± 0.09 | 75.88 Ba ± 0.80 | 0.92 Bb ± 0.03 | 82.79 Aa ± 4.86 | 0.76 ABb ± 0.04 | 82.80 Aa ± 0.63 | 1.09 ABb ± 0.14 | 80.31 Aa ± 0.38 |
| 90 | 0.75 ABc ± 0.01 | 75.51 Aa ± 0.50 | 0.64 Bb ± 0.04 | 65.93 Bb ± 3.31 | 0.62 Bb ± 0.04 | 80.28 Aa ± 1.18 | 0.83 Ac ± 0.04 | 69.69 Bb ± 0.47 | |
| 180 | 1.65 Aa ± 0.15 | 73.10 Ba ± 1.14 | 1.66 Aa ± 0.20 | 79.51 Aa ± 0.24 | 0.72 Bb ± 0.18 | 63.97 Cb ± 1.80 | 1.65 Aab ± 0.11 | 75.74 Bab ± 2.12 | |
| 270 | 0.74 Bc ± 0.01 | 64.76 Bb ± 1.98 | 0.64 Cb ± 0.01 | 77.35 Aa ± 2.03 | 0.82 Ab ± 0.03 | 78.89 Aa ± 1.73 | 0.75 Bc ± 0.02 | 79.15 Aa ± 1.26 | |
| 360 | 1.79 Aa ± 0.17 | 36.25 Cc ± 2.49 | 1.91 Aa ± 0.22 | 62.88 Ab ±4.64 | 1.51 Aa ± 0.16 | 34.30 Cc ± 3.76 | 1.54 Aa ± 0.16 | 48.79 Bc ± 2.34 | |
| Post- gastrointestinal | 28 | 1.42 Ac ± 0.01 | 77.41 Ab ± 4.13 | 1.43 Ab ± 0.03 | 57.52 Cc ± 2.02 | 1.37 Abd ± 0.003 | 63.83 Bc ± 0.31 | 1.46 Ac ± 0.003 | 65.85 Bc ± 0.30 |
| 90 | 1.38 Bc ± 0.02 | 81.93 Ab ± 6.16 | 1.53 Ab ± 0.01 | 59.48 Bc ± 2.94 | 1.08 Dd ± 0.01 | 51.17 Cd ±1.13 | 1.31 Cd ± 0.01 | 59.71 Bc ± 2.25 | |
| 180 | 1.96 Ab ± 0.06 | 92.73 Aa ± 6.25 | 2.02 Aab ± 0.31 | 95.00 Aa ± 1.36 | 1.89 Ab ± 0.02 | 95.82 Aa ± 0.22 | 1.93 Ab ± 0.02 | 93.01 Aa ± 2.23 | |
| 270 | 1.16 Ac ± 0.26 | 82.36 Bb ± 0.17 | 1.76 Ab ± 0.22 | 82.45 Bb ± 0.70 | 1.60 Ac ± 0.13 | 89.50 Ab ± 0.61 | 1.60 Ac ± 0.13 | 83.17 Bb ± 0.68 | |
| 360 | 3.02 Aa ± 0.09 | 39.35 Bc ± 2.17 | 2.73 Aa ± 0.62 | 42.77 Ad ± 3.44 | 2.60 Aa ± 0.16 | 29.54 Ce ± 0.72 | 2.60 Aa ± 0.16 | 28.06 Cd ± 3.78 | |
| After simulated adsorption | 28 | 0.25 Ac ± 0.04 | 68.33 Aa ± 0.30 | 0.24 Ad ± 0.003 | 61.72 Bb ± 0.81 | 0.28 Ac ± 0.05 | 65.05 ABa ± 2.30 | 0.25 Ac ± 0.05 | 57.68 Cb ± 3.04 |
| 90 | 0.78 Aa ± 0.01 | 26.47 Dd ± 1.05 | 0.61 BCb ± 0.04 | 47.79 Cd ± 3.08 | 0.56 Cb ± 0.003 | 51.54 Bb ± 0.47 | 0.68 Ba ± 0.003 | 55.38 Ab ± 1.07 | |
| 180 | 0.76 Aa ± 0.08 | 66.00 Bb ± 0.70 | 0.60 Ac ± 0.02 | 78.10 Aa ± 0.05 | 0.72 Aa ± 0.008 | 65.63 Ba ± 2.50 | 0.68 Aa ± 0.008 | 75.45 Aa ± 2.97 | |
| 270 | 0.88 Aa ± 0.01 | 44.19 Cc ± 1.75 | 0.67 Bab ± 0.07 | 53.75 Ac ± 0.27 | 0.82 Aa ± 0.07 | 39.27 Dc ± 2.42 | 0.67 Ba ± 0.07 | 47.42 Bc ± 1.18 | |
| 360 | 0.50 Bb ± 0.04 | 17.50 Be ± 2.45 | 0.86 Aa ± 0.05 | 17.61 Be ± 0.01 | 0.51 Bb ± 0.05 | 20.68 Bd ± 2.33 | 0.43 Bb ± 0.05 | 27.53 Ad ± 0.01 | |
| Peptide | Mass | Protein | C 1 | LOCK | BB12 | BAUER | A Parameter 2 | Hydrophobicity 3 | Toxicity 4 | Allergenicity Probability 5 | Probability of Being Cell Penetrating 6 | DPP-IViPredictor 7 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WSF | VATPPPPPPPK | 1096.62 | Stress induced phosphoprotein 1 (I3LNG8) | 61.82 | 1.2727 | −0.09 | 0 | 0 | 0.308375 | 414.6, DPPIV | |||
| GEAGPAGPAGPAGPR | 1260.62 | Collagen type I alpha 2 chain (F1SFA7) | 77.72 | 83.14 | 1.2000 | −0.06 | 0 | probably allergen | 0.230702 | 284.93, non-DPPIV | |||
| GDRGEAGPAGPAGPAGPR | 1588.77 | Collagen type I alpha 2 chain (F1SFA7) | 64 | 1.1111 | −0.18 | 0 | probably allergen | 0.272657 | 261.35, non-DPPIV | ||||
| VPVPLPK | 748.484 | Phosphatase and actin regulator (F1S716) | 46.01 | 46.49 | 40.93 | 37.1 | 1.0000 | 0.04 | 0 | 0 | 0.469216 | 383.83, DPPIV | |
| VILPGPAPWG | 1005.56 | PDZ and LIM domain protein 3 (I3L9B6) | 35.71 | 45.95 | 51.76 | 43.16 | 1.0000 | 0.25 | 0 | 0 | 0.179825 | 357.44, DPPIV | |
| NWRPPQPI | 1006.53 | Carbonicanhydrase 3 (Q5S1S4) | 35.83 | 40.39 | 35.24 | 36.74 | 1.0000 | 0.13 | 0 | probably allergen | 0.235725 | 398.71, DPPIV | |
| LILPVGPAGGNQ | 1134.64 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (P80895) | 38.83 | 49.2 | 47.93 | 1.0000 | 0.13 | 0 | probably allergen | 0.166227 | 307.36, DPPIV | ||
| VILPGPAPW | 948.543 | PDZ and LIM domain protein 3 (I3L9B6) | 36.97 | 43.85 | 1.0000 | 0.26 | 0 | 0 | 0.190652 | 381.0, DPPIV | |||
| MLPSLPLL | 882.524 | Basonuclin 1 (F1RIB5) | 41.88 | 46.2 | 1.0000 | 0.25 | 0 | 0 | 0.311738 | 361.29, DPPIV | |||
| HFFNPVPL | 969.507 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial (P00348) | 33.38 | 33.73 | 1.0000 | 0.14 | 0 | 0 | 0.0810493 | 379.57, DPPIV | |||
| LPLVPVPSPGPPAPL | 1449.86 | V-set and immunoglobulin domain containing 10-like (F1RP94) | 32.81 | 31.41 | 1.0000 | 0.16 | 0 | 0 | 0.235179 | 373.36, DPPIV | |||
| VPIPVPLPM | 961.567 | Aldehyde dehydrogenase 6 family member A1 (F1S3H1) | 43.19 | 51.59 | 1.0000 | 0.26 | 0 | 0 | 0.150306 | 425.0, DPPIV | |||
| PQNVILPGPAPWG | 1344.71 | PDZ and LIM domain protein 3 (I3L9B6) | 60.9 | 1.0000 | 0.09 | 0 | 0 | 0.182412 | 359.42, DPPIV | ||||
| APPPPAEVH | 913.465 | Troponin T, fast skeletal muscle (Q75NG9) | 31.65 | 1.0000 | −0.03 | 0 | probably allergen | 0.121088 | 401.0, DPPIV | ||||
| GLPPPGLT | 750.427 | Uncharacterized protein (F1SFN5) | 40.66 | 1.0000 | 0.12 | 0 | probably allergen | 0.242026 | 374.14, DPPIV | ||||
| PLALAGPPPP | 928.538 | Androgen receptor (Q9GKL7) | 36.81 | 1.0000 | 0.14 | 0 | 0 | 0.259959 | 395.89, DPPIV | ||||
| EPVPLAHPLP | 1068.59 | Lactamase beta-like 1 (F1STY0) | 33.58 | 1.0000 | 0.06 | 0 | 0 | 0.176067 | 394.89, DPPIV | ||||
| PQNVILPGPAPW | 1287.69 | PDZ and LIM domain protein 3 (I3L9B6) | 39.14 | 1.0000 | 0.08 | 0 | 0 | 0.188348 | 376.73, DPPIV | ||||
| PVVPPLIPPK | 1055.67 | Troponin T, slow skeletal muscle (Q75ZZ6) | 62.9 | 1.0000 | 0.09 | 0 | 0 | 0.24149 | 388.0, DPPIV | ||||
| AVSPGLAGPATK | 1067.59 | Membrane integral NOTCH2 associated receptor 1 (F1RKQ2) | 41.94 | 1.0000 | 0.04 | 0 | 0 | 0.300649 | 259.45, non-DPPIV | ||||
| SSF | GPAGPAGPAGPR | 1003.52 | Collagen type I alpha 2 chain (F1SFA7) | 56.91 | 1.3333 | −0.05 | 0 | probable allergen | 0.26505 | 302.55, DPPIV | |||
| AGPAGPAGPAGPR | 1074.55 | Collagen type I alpha 2 chain (F1SFA7) | 56.12 | 58.46 | 1.3077 | −0.03 | 0 | probable allergen | 0.288459 | 295.33, DPPIV | |||
| PAGPAGPAGPR | 946.498 | Collagen type I alpha 2 chain (F1SFA7) | 49.15 | 66.96 | 1.1818 | −0.07 | 0 | probable allergen | 0.279523 | 315.9, DPPIV | |||
| IPAPPGKP | 775.459 | Guanine nucleotide exchange factor VAV2 isoform 2 (F1S039) | 41.85 | 1.0000 | −0.03 | 0 | 0 | 0.159305 | 362.0, DPPIV | ||||
| APPPPAEV | 776.406 | Troponin T, fast skeletal muscle (Q75NG9) | 44.25 | 1.0000 | 0.02 | 0 | probable allergen | 0.146887 | 417.0, DPPIV | ||||
| KLPPLPL | 776.51 | Translocase of outer mitochondrial membrane 40 (F1RM44) | 30.16 | 1.0000 | 0.04 | 0 | 0 | 0.550444 | 380.5, DPPIV | ||||
| VPLPVPVPI | 929.59 | Retinoicacidinduced 2 (F1SQQ4) | 33.14 | 1.0000 | 0.29 | 0 | 0 | 0.188433 | 412.88, DPPIV | ||||
| GPPGPPGKP | 802.43 | Uncharacterized protein (I3L8B2) | 71.03 | 1.0000 | −0.11 | 0 | 0 | 0.204097 | 363.38, DPPIV | ||||
| RIPIIP | 707.469 | RING-type E3 ubiquitin transferase (F1RFJ1) | 30.97 | 1.0000 | 0.05 | 0 | probable allergen | 0.100824 | 346.2, DPPIV | ||||
| RLPLLP | 707.469 | Kelch-like family member 4 (F1S1P2) | 30.97 | 1.0000 | −0.05 | 0 | probable allergen | 0.73246 | 377.4, DPPIV | ||||
| SKRLPLP | 809.512 | Uncharacterized protein (I3LRP9) | 34.66 | 1.0000 | −0.31 | 0 | probable allergen | 0.609697 | 277.17, non-DPPIV | ||||
| WVGLPPLPSA | 1035.57 | Bardet–Biedl syndrome 5 protein homolog (F1S1V8) | 46.01 | 1.0000 | 0.19 | 0 | 0 | 0.239102 | 350.78, DPPIV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kęska, P.; Stadnik, J. Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Dry-Cured Pork Loin during Aging and Gastrointestinal Digestion. Nutrients 2022, 14, 770. https://doi.org/10.3390/nu14040770
Kęska P, Stadnik J. Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Dry-Cured Pork Loin during Aging and Gastrointestinal Digestion. Nutrients. 2022; 14(4):770. https://doi.org/10.3390/nu14040770
Chicago/Turabian StyleKęska, Paulina, and Joanna Stadnik. 2022. "Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Dry-Cured Pork Loin during Aging and Gastrointestinal Digestion" Nutrients 14, no. 4: 770. https://doi.org/10.3390/nu14040770
APA StyleKęska, P., & Stadnik, J. (2022). Dipeptidyl Peptidase IV Inhibitory Peptides Generated in Dry-Cured Pork Loin during Aging and Gastrointestinal Digestion. Nutrients, 14(4), 770. https://doi.org/10.3390/nu14040770