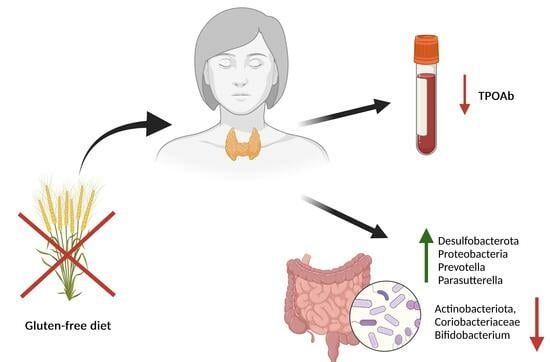
Gluten-Free Diet Alters the Gut Microbiome in Women with Autoimmune Thyroiditis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Study Design
2.3. Compounding of Capsules and Their Quality Assessment
2.4. Laboratory Tests
2.5. Microbiome Testing and Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
3.1. Microbiome Samples
3.2. Changes in Microbiome
3.2.1. Changes in Microbiome after 4 Weeks of Gluten-Free Diet
3.2.2. Differences between the Gluten and Placebo Groups in the Microbiome at the Time of Allocation (T1)
3.2.3. Changes in Microbiome within the Gluten and Placebo Groups after 4 Weeks of Capsule Intake
3.2.4. Differences in Microbiome between the Gluten and Placebo Groups after Whole Intervention
3.3. Blood Parameter Analysis
3.4. Changes in Blood Parameters
3.4.1. Changes in Blood Parameters after 4 Weeks of a Gluten-Free Diet
3.4.2. Differences between the Gluten and Placebo Groups in the Blood Parameters at the Time of Allocation
3.4.3. Changes in Blood Parameters within the Gluten and Placebo Groups after 4 Weeks of Capsule Intake
3.4.4. Differences in Blood Parameters between the Gluten and Placebo Groups after Whole Intervention
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGrogan, A.; Seaman, H.E.; Wright, J.W.; De Vries, C.S. The incidence of autoimmune thyroid disease: A systematic review of the literature. Clin. Endocrinol. 2008, 69, 687–696. [Google Scholar] [CrossRef]
- Caturegli, P.; Kimura, H.; Rocchi, R.; Rose, N.R. Autoimmune thyroid diseases. Curr. Opin. Rheumatol. 2007, 19, 44–48. [Google Scholar] [CrossRef]
- McLeod, D.S.A.; Cooper, D.S. The incidence and prevalence of thyroid autoimmunity. Endocrine 2012, 42, 252–265. [Google Scholar] [CrossRef]
- Vanderpump, M.P.J. The epidemiology of thyroid disease. Br. Med. Bull. 2011, 99, 39–51. [Google Scholar] [CrossRef]
- Proal, A.D.; Albert, P.J.; Marshall, T.G. The human microbiome and autoimmunity. Curr. Opin. Rheumatol. 2013, 25, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, H.M.; Mohammad, I.S.; Guo, H.; Shahzad, M.; Hou, Y.J.; Ma, C.; Naseem, Z.; Wu, X.; Shi, P.; Xu, J. Molecular estimation of alteration in intestinal microbial composition in Hashimoto’s thyroiditis patients. Biomed. Pharmacother. 2017, 95, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut Microbiota 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Krysiak, R.; Szkróbka, W.; Okopień, B. The Effect of Gluten-Free Diet on Thyroid Autoimmunity in Drug-Naïve Women with Hashimoto’s Thyroiditis: A Pilot Study. Exp. Clin. Endocrinol. Diabetes 2019, 127, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Pobłocki, J.; Pańka, T.; Szczuko, M.; Telesiński, A.; Syrenicz, A. Whether a gluten-free diet should be recommended in chronic autoimmune thyroiditis or not?—A 12-month follow-up. J. Clin. Med. 2021, 10, 3240. [Google Scholar] [CrossRef]
- Ch’ng, C.L.; Jones, M.K.; Kingham, J.G.C. Celiac disease and autoimmune thyroid disease. Clin. Med. Res. 2007, 5, 184–192. [Google Scholar] [CrossRef]
- Roy, A.; Laszkowska, M.; Sundström, J.; Lebwohl, B.; Green, P.H.; Kämpe, O.; Ludvigsson, J.F. Prevalence of Celiac Disease in Patients with Autoimmune Thyroid Disease: A Meta-Analysis. Thyroid 2016, 26, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Bai, J.C.; Bonaz, B.; Bouma, G.; Calabrò, A.; Carroccio, A.; Castillejo, G.; Ciacci, C.; Cristofori, F.; Dolinsek, J.; et al. Non-celiac gluten sensitivity: The new frontier of gluten related disorders. Nutrients 2013, 5, 3839–3853. [Google Scholar] [CrossRef]
- Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult humans. Gut Microbes 2010, 1, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Caminero, A.; Vivas, S.; Ruiz de Morales, J.M.; Sáenz de Miera, L.E.; Rodríguez-Aparicio, L.B.; Casqueiro, J. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie 2012, 94, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Di Sabatino, A.; Volta, U.; Salvatore, C.; Biancheri, P.; Caio, G.; De Giorgio, R.; Di Stefano, M.; Corazza, G.R. Small Amounts of Gluten in Subjects with Suspected Nonceliac Gluten Sensitivity: A Randomized, Double-Blind, Placebo-Controlled, Cross-Over Trial. Clin. Gastroenterol. Hepatol. 2015, 13, 1604–1612.e3. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, M.; Tomczak, E. The need to report effect size estimates revisited. An overview of some recommended measures of effect size. Trends Sport Sci. 2014, 1, 19–25. Available online: https://wbc.poznan.pl/dlibra/publication/413565/edition/325867/content (accessed on 9 February 2023).
- Cohen, J. A power primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Correlation between body mass index and faecal microbiota from children. Clin. Microbiol. Infect. 2016, 22, 258.e1–258.e8. [Google Scholar] [CrossRef]
- Patil, D.P.; Dhotre, D.P.; Chavan, S.G.; Sultan, A.; Jain, D.S.; Lanjekar, V.B.; Gangawani, J.; Shah, P.S.; Todkar, J.S.; Shah, S.; et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J. Biosci. 2012, 37, 647–657. [Google Scholar] [CrossRef]
- El-Zawawy, H.T.; Ahmed, S.M.; El-Attar, E.A.; Ahmed, A.A.; Roshdy, Y.S.; Header, D.A. Study of gut microbiome in Egyptian patients with autoimmune thyroid diseases. Int. J. Clin. Pract. 2021, 75, e14038. [Google Scholar] [CrossRef] [PubMed]
- Cayres, L.C.d.F.; de Salis, L.V.V.; Rodrigues, G.S.P.; Lengert, A.v.H.; Biondi, A.P.C.; Sargentini, L.D.B.; Brisotti, J.L.; Gomes, E.; de Oliveira, G.L.V. Detection of Alterations in the Gut Microbiota and Intestinal Permeability in Patients with Hashimoto Thyroiditis. Front. Immunol. 2021, 12, 579140. [Google Scholar] [CrossRef]
- Zhao, F.; Feng, J.; Li, J.; Zhao, L.; Liu, Y.; Chen, H.; Jin, Y.; Zhu, B.; Wei, Y. Alterations of the gut microbiota in Hashimoto’s thyroiditis patients. Thyroid 2018, 28, 175–186. [Google Scholar] [CrossRef]
- Xu, Z.; Lian, C.; Pan, L.; Lai, W.; Zhang, F.; Peng, L.; Zhou, S.; Zhao, G.; Yang, X.; Zhang, G.; et al. N-acetyl-L-leucine protects MPTP-treated Parkinson’s disease mouse models by suppressing Desulfobacterota via the gut-brain axis. Brain Res. Bull. 2023, 202, 110729. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wan, Z.; Zhang, Y.; Wang, T.; Xue, Y.; Peng, Q. Composition and diversity of gut microbiota in diabetic retinopathy. Front. Microbiol. 2022, 13, 926926. [Google Scholar] [CrossRef]
- Amar, J.; Lange, C.; Payros, G.; Garret, C.; Chabo, C.; Lantieri, O.; Courtney, M.; Marre, M.; Charles, M.A.; Balkau, B.; et al. Blood Microbiota Dysbiosis Is Associated with the Onset of Cardiovascular Events in a Large General Population: The D.E.S.I.R. Study. PLoS ONE 2013, 8, e54461. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Gophna, U.; Sommerfeld, K.; Gophna, S.; Doolittle, W.F.; Van Zanten, S.J.O.V. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006, 44, 4136–4141. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, W.; Kosik, R.O.; Song, Y.; Luo, Q.; Qiao, T.; Tong, J.; Liu, S.; Deng, C.; Qin, S.; et al. Gut microbiota changes and its potential relations with thyroid carcinoma. J. Adv. Res. 2022, 35, 61–70. [Google Scholar] [CrossRef]
- Yang, M.; Sun, B.; Li, J.; Yang, B.; Xu, J.; Zhou, X.; Yu, J.; Zhang, X.; Zhang, Q.; Zhou, S.; et al. Alteration of the intestinal flora may participate in the development of graves’ disease: A study conducted among the han population in Southwest China. Endocr. Connect. 2019, 8, 822–828. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Precup, G.; Vodnar, D.-C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef]
- Ortega-Santos, C.P.; Whisner, C.M. The Key to Successful Weight Loss on a High-Fiber Diet May Be in Gut Microbiome Prevotella Abundance. J. Nutr. 2019, 149, 2083–2084. [Google Scholar] [CrossRef]
- Eriksen, A.K.; Brunius, C.; Mazidi, M.; Hellström, P.M.; Risérus, U.; Iversen, K.N.; Fristedt, R.; Sun, L.; Huang, Y.; Nørskov, N.P.; et al. Effects of whole-grain wheat, rye, and lignan supplementation on cardiometabolic risk factors in men with metabolic syndrome: A randomized crossover trial. Am. J. Clin. Nutr. 2020, 111, 864–876. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Ibrahim, M.; Subramanian, A.; Anishetty, S. Comparative pan genome analysis of oral Prevotella species implicated in periodontitis. Funct. Integr. Genom. 2017, 17, 513–536. [Google Scholar] [CrossRef]
- Si, J.; You, H.J.; Yu, J.; Sung, J.; Ko, G. Prevotella as a Hub for Vaginal Microbiota under the Influence of Host Genetics and Their Association with Obesity. Cell Host Microbe 2017, 21, 97–105. [Google Scholar] [CrossRef]
- Armstrong, A.J.S.; Shaffer, M.; Nusbacher, N.M.; Griesmer, C.; Fiorillo, S.; Schneider, J.M.; Neff, C.P.; Li, S.X.; Fontenot, A.P.; Campbell, T.; et al. Correction to: An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2020, 8, 50. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H.; Wu, S.; Lu, N.; Wang, Y.; Liu, H.; Dong, L.; Liu, T.; Shen, X. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018, 33, 1844–1852. [Google Scholar] [CrossRef]
- Henneke, L.; Schlicht, K.; Andreani, N.A.; Hollstein, T.; Demetrowitsch, T.; Knappe, C.; Hartmann, K.; Jensen-Kroll, J.; Rohmann, N.; Pohlschneider, D.; et al. A dietary carbohydrate–gut Parasutterella–human fatty acid biosynthesis metabolic axis in obesity and type 2 diabetes. Gut Microbes 2022, 14, 2057778. [Google Scholar] [CrossRef] [PubMed]
- Kreutzer, C.; Peters, S.; Schulte, D.M.; Fangmann, D.; Türk, K.; Wolff, S.; van Eimeren, T.; Ahrens, M.; Beckmann, J.; Schafmayer, C.; et al. Hypothalamic inflammation in human obesity is mediated by environmental and genetic factors. Diabetes 2017, 66, 2407–2415. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Liu, M.; Fujimura, K.E.; Lyalina, S.; Nagarkar, D.R.; Charbit, B.; Bergstedt, J.; Patin, E.; Harrison, O.J.; Quintana-Murci, L.; et al. Gut microbiome stability and dynamics in healthy donors and patients with non-gastrointestinal cancers. J. Exp. Med. 2020, 218, e20200606. [Google Scholar] [CrossRef] [PubMed]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Liu, B.; Ding, Z.; Xiong, J.; Heng, X.; Wang, H.; Chu, W. Gut Microbiota and Inflammatory Cytokine Changes in Patients with Ankylosing Spondylitis. BioMed Res. Int. 2022, 2022, 1005111. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xiao, G.; Yao, Y.; Guo, S.; Lu, K.; Sheng, Z. The Role of Bifidobacteria in Gut Barrier Function After Thermal Injury in Rats. J. Trauma Inj. Infect. Crit. Care 2006, 61, 650–657. [Google Scholar] [CrossRef]
- Griffiths, E.A.; Duffy, L.C.; Schanbacher, F.L.; Qiao, H.; Dryja, D.; Leavens, A.; Rossman, J.; Rich, G.; Dirienzo, D.; Ogra, P.L. In vivo effects of bifidobacteria and lactoferrin on gut endotoxin concentration and mucosal immunity in Balb/c mice. Dig. Dis. Sci. 2004, 49, 579–589. [Google Scholar] [CrossRef] [PubMed]
- López, P.; Gueimonde, M.; Margolles, A.; Suárez, A. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 2010, 138, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ni, J.-J.; Han, B.-X.; Yan, S.-S.; Wei, X.-T.; Feng, G.-J.; Zhang, H.; Zhang, L.; Li, B.; Pei, Y.-F. Causal Relationship Between Gut Microbiota and Autoimmune Diseases: A Two-Sample Mendelian Randomization Study. Front. Immunol. 2022, 12, 746998. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zou, L.; Nie, C.; Qin, Y.; Tong, X.; Wang, J.; Yang, H.; Xu, X.; Jin, X.; Xiao, L.; et al. Mendelian randomization analyses reveal causal relationships between the human microbiome and longevity. Sci. Rep. 2023, 13, 5127. [Google Scholar] [CrossRef]
- Jin, Q.; Ren, F.; Dai, D.; Sun, N.; Qian, Y.; Song, P. The causality between intestinal flora and allergic diseases: Insights from a bi-directional two-sample Mendelian randomization analysis. Front. Immunol. 2023, 14, 1121273. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients with vs without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef]
- Kim, M.-H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.-L.; Kim, H.-N. Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A.; et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013, 7, 269–280. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.B.S.; Roager, H.M.; Søndertoft, N.B.; Gøbel, R.J.; Kristensen, M.; Vallès-Colomer, M.; Vieira-Silva, S.; Ibrügger, S.; Lind, M.V.; Mærkedahl, R.B.; et al. A low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults. Nat. Commun. 2018, 9, 4630. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Collado, M.C.; Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 2009, 102, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, J.; Starchl, C.; Berisha, A.T.; Amrein, K. Thyroid-gut-axis: How does the microbiota influence thyroid function? Nutrients 2020, 12, 1769. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Zafeiropoulou, K.; Nichols, B.; Mackinder, M.; Biskou, O.; Rizou, E.; Karanikolou, A.; Clark, C.; Buchanan, E.; Cardigan, T.; Duncan, H.; et al. Alterations in Intestinal Microbiota of Children with Celiac Disease at the Time of Diagnosis and on a Gluten-free Diet. Gastroenterology 2020, 159, 2039–2051.e20. [Google Scholar] [CrossRef]



| Taxonomic Level | Bacterial Taxa |
|---|---|
| Phylum | Actinobacteriota, Bacteroidota, Cyanobacteria, Desulfobacterota, Firmicutes, Fusobacteriota, Proteobacteria, Verrucomicrobiota |
| Family | Prevotellaceae, Veillonellaceae, Clostridiaceae, Coriobacteriaceae, Enterobacteriaceae, Victivallaceae |
| Genus | Bifidobacterium, Lactobacillus, Dialister, Prevotella, Alistipes, Escherichia-Shigella, Parasutterella, Slackia |
| Species | Escherichia coli, Akkermansia (uncultured bacterium) |
| Bacteria | npairs | Bacterial Abundance, Mdn (Q1–Q3) | p | r | |
|---|---|---|---|---|---|
| Baseline (T0) | After 4 Weeks (T1) | ||||
| Phylum | |||||
| Actinobacteriota | 30 | 3199.0 (2165.0–5285.0) | 2962.5 (1697.3–3626.8) | 0.038 | 0.38 |
| Bacteroidota | 30 | 16,684.5 (11,372.5–18,349.8) | 16,112.5 (14,660.0–18,170.0) | 0.700 | 0.07 |
| Cyanobacteria | 30 | 3.0 (0–632.0) | 58.5 (99.5–257.5) | 0.639 | 0.07 |
| Desulfobacterota | 30 | 213.0 (155.0–333.0) | 294.0 (159.8–454.8) | 0.021 | 0.41 |
| Firmicutes | 30 | 34,854.5 (31,463.0–39,079.3) | 37,647.5 (30,931.5–40,956.0) | 0.221 | 0.23 |
| Fusobacteriota | 30 | 0 (0–0) | 0 (0–0) | 0.889 | 0.11 |
| Proteobacteria | 30 | 1667.0 (962.0–2440.0) | 2408.5 (1402.0–4711.8) | 0.015 | 0.44 |
| Verrucomicrobiota | 30 | 507.0 (194.5–1761.8) | 734 (307.3–2109.8) | 0.761 | 0.06 |
| Family | |||||
| Prevotellaceae | 30 | 1375.0 (17.5–4595) | 1456.5 (2.0–3000.8) | 0.105 | 0.30 |
| Veillonellaceae | 30 | 180.0 (8.5–1017.0) | 95.0 (9.8–754.8) | 0.146 | 0.22 |
| Clostridiaceae | 30 | 89.5 (38.8–245.0) | 78.0 (15.8–565.8) | 0.728 | 0.02 |
| Coriobacteriaceae | 30 | 967.5 (578.0–1992.5) | 731.0 (288.3–1340.3) | 0.003 | 0.52 |
| Enterobacteriaceae | 30 | 129.0 (44.0–337.5) | 119.5 (23.0–518.8) | 0.746 | 0.06 |
| Victivallaceae | 30 | 17.5 (0–38.8) | 21.5 (0–48.5) | 0.334 | 0.16 |
| Genus | |||||
| Bifidobacterium | 30 | 1238.5 (565.8–2983.0) | 672.5 (309.3–1643.3) | 0.011 | 0.46 |
| Lactobacillus | 30 | 4.5 (0–20.8) | 2.5 (0–15.3) | 0.158 | 0.30 |
| Dialister | 30 | 66.5 (0–999.0) | 10.5 (0–667.0) | 0.194 | 0.22 |
| Prevotella | 30 | 66.0 (9.3–3402.0) | 93.0 (2.0–1610.3) | 0.044 | 0.37 |
| Alistipes | 30 | 1249.5 (716.8–2385.5) | 1332 (986.0–2608.0) | 0.727 | 0.07 |
| Escherichia-Shigella | 30 | 78.0 (26.0–327.0) | 99.5 (16.3–482.0) | 0.729 | 0.07 |
| Parasutterella | 30 | 43.5 (14.3–153.8) | 72.5 (18.3–266.3) | 0.008 | 0.49 |
| Slackia | 30 | 20.5 (0–199.3 ) | 15.5 (0–182.8) | 0.237 | 0.177 |
| Species | |||||
| Escherichia coli | 30 | 78.0 (26.0–327.0) | 99.5 (16.3–482.0) | 0.729 | 0.07 |
| Akkermansia | 30 | 409.5 (126.5–1256.0) | 358.0 (207.5–1913.5) | 1.000 | 0.02 |
| Bacteria | npairs | Group | Bacterial Abundance, Mdn (Q1–Q3) | p | r | |
|---|---|---|---|---|---|---|
| After 4 Weeks (T1) | After 8 Weeks (T2) | |||||
| Phylum | ||||||
| Actinobacteriota | 14 | gluten | 2622.5 (1964.2–3224.0) | 2736.0 (1767.5–4918.2) | 0.502 | 0.19 |
| 15 | placebo | 2728.0 (1228.5–3531.5) | 3412.0 (1565.0–4603.0) | 0.030 | 0.56 | |
| Bacteroidota | 14 | gluten | 15,816.5 (14,591.0–18,856.5) | 16,310.0 (11,928.0–19,632.5) | 0.542 | 0.18 |
| 15 | placebo | 16,188.0 (15,573.0–17,939.0) | 19,942.0 (15,962.0–21,380.0) | 0.015 | 0.62 | |
| Cyanobacteria | 14 | gluten | 186.5 (1.5–654.0) | 111.5 (313.6–523.0) | 1.000 | 0.01 |
| 15 | placebo | 4.0 (0–151.5 ) | 0 (0–144.0) | 0.906 | 0.16 | |
| Desulfobacterota | 14 | gluten | 281.5 (183.5–496.5) | 346.0 (187.5–517.2) | 0.889 | 0.05 |
| 15 | placebo | 335.0 (155.5–383.5) | 266.0 (157.0–369.5) | 0.303 | 0.28 | |
| Firmicutes | 14 | gluten | 37,266.5 (33,142.2–40,886.0) | 35,924.0 (30,814.8–41,329.8) | 0.583 | 0.16 |
| 15 | placebo | 35,393.0 (30,649.0–40,745.5) | 35,828.0 (33,275.0–42,841.5) | 0.188 | 0.35 | |
| Fusobacteriota | 14 | gluten | 0 (0–0) | 0 (0–0) | 0.584 | 0.04 |
| 15 | placebo | 0 (0–2.5) | 0 (0–0) | 0.178 | 0.36 | |
| Proteobacteria | 14 | gluten | 3002.0 (1416.2–4748.5) | 3488.5 (2149.5–5852.5) | 0.326 | 0.28 |
| 15 | placebo | 2462.0 (1749.5–4070.5) | 2496.0 (1655.0–4494.5) | 0.599 | 0.15 | |
| Verrucomicrobiota | 14 | gluten | 500.5 (274.0–1210.0) | 471.0 (171.0–2791.5) | 0.235 | 0.34 |
| 15 | placebo | 1273.0 (446.5–2138.0) | 798.0 (356.0–1717.5) | 0.421 | 0.22 | |
| Family | ||||||
| Prevotellaceae | 14 | gluten | 2349.5 (99.0–3476.2) | 1682.0 (124.2–3387.2) | 0.889 | 0.07 |
| 15 | placebo | 745.0 (2.0–2363.0) | 851.0 (0.0–4747.5) | 0.255 | 0.22 | |
| Veillonellaceae | 14 | gluten | 158.0 (9.8–961.0) | 30.0 (0.0–480.0) | 0.056 | 0.55 |
| 15 | placebo | 74.0 (17.0–390.0) | 109.0 (9.0–671.0) | 0.889 | 0.04 | |
| Clostridiaceae | 14 | gluten | 162.0 (35.0–1361.2) | 192.0 (108.0–457.0) | 0.328 | 0.25 |
| 15 | placebo | 68.0 (11.0–459.5) | 202 (35.5–695.5) | 0.532 | 0.17 | |
| Coriobacteriaceae | 14 | gluten | 1010.5 (415.5–1507.5) | 1726.5 (8326.8–2225.0) | 0.241 | 0.33 |
| 15 | placebo | 654.0 (300.0–1169.5) | 994.0 (454.5–1928.0) | 0.117 | 0.42 | |
| Enterobacteriaceae | 14 | gluten | 109.0 (19.2–648.2) | 503.0 (54.2–2269.5) | 0.780 | 0.08 |
| 15 | placebo | 132.0 (35.0–317.0) | 156.0 (47.5–427.0) | 0.244 | 0.31 | |
| Victivallaceae | 14 | gluten | 23.5 (0–48.5) | 16.5 (0–67.2) | 0.906 | 0.02 |
| 15 | placebo | 21.0 (0–65.0) | 20.0 (0–98.5) | 0.359 | 0.14 | |
| Genus | ||||||
| Bifidobacterium | 14 | gluten | 731.0 (400.2–1523.2) | 882.5 (560.2–1899.8) | 0.358 | 0.26 |
| 15 | placebo | 1154.7 (229.5–1292.0) | 801.0 (470.5–1623.0) | 0.184 | 0.33 | |
| Lactobacillus | 14 | gluten | 5.0 (0–41.5) | 1.5 (0–23.0) | 1.000 | 0.06 |
| 15 | placebo | 2.0 (0–8.5) | 9.0 (0–20.5) | 0.221 | 0.34 | |
| Dialister | 14 | gluten | 7.5 (0–946.2) | 0 (0–480.8) | 0.236 | 0.311 |
| 15 | placebo | 52.0 (0–390.0) | 0 (0–660.5) | 0.722 | 0.19 | |
| Prevotella | 14 | gluten | 149.5 (6.5–1783.2) | 100.5 (6.2–1946.5) | 0.780 | 0.10 |
| 15 | placebo | 6.0 (1.0–1585.5) | 25.0 (0.0–3935.0) | 0.197 | 0.31 | |
| Alistipes | 14 | gluten | 1302.8 (990.2–2243.0) | 1836.5 (904.8–2040.2) | 0.217 | 0.34 |
| 15 | placebo | 1207.0 (892.5–2973.0) | 1125.0 (732.5–2637.0) | 0.679 | 0.12 | |
| Escherichia-Shigella | 14 | gluten | 41.0 (3.8–641.0) | 489.0 (40.8–2268.8) | 0.784 | 0.10 |
| 15 | placebo | 132.0 (35.0–317.0) | 156.0 (21.5–392.5) | 0.244 | 0.31 | |
| Parasutterella | 14 | gluten | 91.5 (22.2–334.0) | 117.5 (34.8–280.5) | 0.660 | 0.13 |
| 15 | placebo | 80.0 (30.0–203.5) | 68.0 (18.5–230.5) | 0.315 | 0.30 | |
| Slackia | 14 | gluten | 21.5 (0–498.8) | 25.5 (63.1–229.8) | 0.326 | 0.24 |
| 15 | placebo | 16.0 (47.7–160.5) | 9.0 (53.1–242.0) | 0.760 | 0.02 | |
| Species | ||||||
| Escherichia coli | 14 | gluten | 41.0 (3.8–641.0) | 489.0 (40.8–2094.8) | 0.724 | 0.12 |
| 15 | placebo | 132.0 (35.0–317.0) | 156.0 (21.5–392.%) | 0.244 | 0.31 | |
| Akkermansia | 14 | gluten | 318.0 (272.8–1157.0) | 338.0 (50.2–2727.0) | 0.367 | 0.24 |
| 15 | placebo | 852.0 (176.0–2105.5) | 488.0 (711.1–1608.5) | 0.530 | 0.18 | |
| Parameter | npairs | Distribution of Blood Parameters, M (SD) 1 | p | d2 | |
|---|---|---|---|---|---|
| Baseline (T0) | After 4 Weeks (T1) | ||||
| TSH [µIU/mL] | 30 | 2.15 (1.2) | 2.14 (1.08) | 0.969 | 0.01 |
| FT4 [ng/dL] | 30 | 1.32 (0.20) | 1.31 (0.20) | 0.551 | 0.11 |
| FT3 [pg/mL] | 30 | 2.88 (0.32) | 2.86 (0.33) | 0.675 | 0.08 |
| Anti-TPO [IU/mL] | 30 | 152.84 (144.00) | 145.31 (138.3) | 0.154 | 0.27 |
| Anti-TG [IU/mL] | 30 | 244.39 (187.8) | 218.99 (170.92) | 0.142 | 0.28 |
| Glucose [mg/dL] | 30 | 87.47 (6.28) | 88.07 (6.92) | 0.546 | –0.11 |
| ESR [mm/h] | 30 | 3.0 (2.0–8.0) | 3.0 (2.0–8.0) | 0.680 | 0.08 |
| CRP [mg/L] | 30 | 0.50 (0.50–0.78) | 0.50 (0.50–1.10) | 0.407 | 0.15 |
| Parameter | Group | Distribution of Blood Parameters, M (SD) 1 | p | d2 | |
|---|---|---|---|---|---|
| After 4 Weeks (T1) | After 8 Weeks (T2) | ||||
| TSH [µIU/mL] | gluten | 2.08 (0.87) | 2.08 (0.78) | 0.685 | 0.12 |
| placebo | 2.20 (1.27) | 2.06 (1.11) | 0.626 | 0.13 | |
| FT4 [ng/dL] | gluten | 1.27 (0.14) | 1.32 (0.19) | 0.175 | −0.40 |
| placebo | 1.34 (0.25) | 1.34 (0.20) | 0.978 | −0.01 | |
| FT3 [pg/mL] | gluten | 2.81 (0.34) | 2.79 (0.34) | 0.769 | 0.08 |
| placebo | 2.90 (0.33) | 2.89 (0.30) | 0.882 | 0.04 | |
| Anti-TPO [IU/mL] | gluten | 99.90 (88.0) | 86.17 (69.98) | 0.067 | 0.56 |
| placebo | 190.71 (165.70) | 184.56 (166.99) | 0.024 | 0.66 | |
| Anti-TG [IU/mL] | gluten | 215.05 (187.42) | 236.42 (199.06) | 0.722 | 0.10 |
| placebo | 222.93 (159.23) | 203.81 (153.56) | 0.110 | 0.44 | |
| Glucose [mg/dL] | gluten | 85.13 (6.16) | 85.62 (4.96) | 0.561 | −0.17 |
| placebo | 91.00 (6.55) | 90.67 (4.40) | 0.711 | 0.10 | |
| ESR [mm/h] | gluten | 3.00 (2.50–8.00) | 3.00 (3.00–9.00) | 0.605 | 0.23 |
| placebo | 3.00 (2.00–7.70) | 3.00 (2.00–5.50) | 0.313 | 0.20 | |
| CRP [mg/L] | gluten | 0.60 (0.50–1.50) | 0.50 (0.50–0.90) | 0.121 | 0.42 |
| placebo | 0.50 (0.50–0.70) | 0.50 (0.50–0.55) | 0.353 | 0.16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodziewicz, A.; Szewczyk, A.; Bryl, E. Gluten-Free Diet Alters the Gut Microbiome in Women with Autoimmune Thyroiditis. Nutrients 2024, 16, 685. https://doi.org/10.3390/nu16050685
Rodziewicz A, Szewczyk A, Bryl E. Gluten-Free Diet Alters the Gut Microbiome in Women with Autoimmune Thyroiditis. Nutrients. 2024; 16(5):685. https://doi.org/10.3390/nu16050685
Chicago/Turabian StyleRodziewicz, Aleksandra, Adrian Szewczyk, and Ewa Bryl. 2024. "Gluten-Free Diet Alters the Gut Microbiome in Women with Autoimmune Thyroiditis" Nutrients 16, no. 5: 685. https://doi.org/10.3390/nu16050685
APA StyleRodziewicz, A., Szewczyk, A., & Bryl, E. (2024). Gluten-Free Diet Alters the Gut Microbiome in Women with Autoimmune Thyroiditis. Nutrients, 16(5), 685. https://doi.org/10.3390/nu16050685