When Food Meets Man: the Contribution of Epigenetics to Health
Abstract
:1. Introduction
2. Basic Concepts in Epigenetics

2.1. The Histone Code
2.2. Chromatin Remodeling Enzymes
2.3. Histone Deacetylases
2.4. Beyond Histones
| Class | Protein domains | Members |
|---|---|---|
| Class I | Deacetylase catalytic domain | HDAC1 |
| Phosphorylation sites (serine residues) at C terminus | HDAC2 | |
| HDAC3 | ||
| HDAC8 | ||
| Class IIa | Deacetylase catalytic domain | HDAC4 |
| Phosphorylation sites (serine residues) at N terminus | HDAC5 | |
| Myocyte enhancer factor binding sites | HDAC7 | |
| Binding sites for 14-3-3 chaperone protein | HDAC9 | |
| Class IIb | Deacetylase catalytic domain | HDAC6 |
| Zinc finger domain or leucine rich region | HDAC10 | |
| Class IV | Deacetylase catalytic domain | HDAC11 |
3. Epigenetic Regulation of Metabolic Pathways
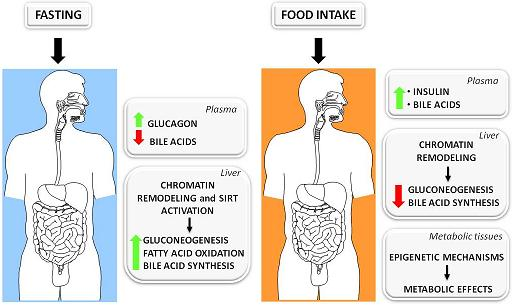
3.1. The Fed-Fasting Cycle: the Role of Bile Acids


3.2. Sirtuins: Key Players in the Fed-Fasting Cycle and Calorie Restriction

3.3. Bile Acid-Induced Post-Translational Modifications of Histones and Chromatin Remodeling

4. Targeting Chromatin and Chromatin-Associated Factors: New Opportunities for Handling Metabolic Disorders
4.1. Histone Deacetylase Inhibitors
4.2. Effects of Histone Deacetylase Inhibitors on Cholesterol Metabolism
4.3. Effects of Histone Deacetylase Inhibitors on Energy Metabolism
4.4. Effects of SIRT Activators on Energy Metabolism
5. Final Considerations
Acknowledgements
References
- Cheung, P.; Allis, C.D.; Sassone-Corsi, P. Signaling to chromatin through histone modifications. Cell 2000, 103, 263–271. [Google Scholar]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar]
- Zhang, L.; Eugeni, E.E.; Parthun, M.R.; Freitas, M.A. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 2003, 112, 77–86. [Google Scholar]
- Kouzarides, T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar]
- Brown, C.E.; Lechner, T.; Howe, L.; Workman, J.L. The many HATs of transcription coactivators. Trends Biochem. Sci. 2000, 25, 15–19. [Google Scholar]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar]
- McKinsey, T.A.; Zhang, C.L.; Lu, J.; Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000, 408, 106–111. [Google Scholar]
- Lu, J.; McKinsey, T.A.; Nicol, R.L.; Olson, E.N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. U S A 2000, 97, 4070–4075. [Google Scholar]
- Passier, R.; Zeng, H.; Frey, N.; Naya, F.J.; Nicol, R.L.; McKinsey, T.A.; Overbeek, P.; Richardson, J.A.; Grant, S.R.; Olson, E.N. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 2000, 105, 1395–1406. [Google Scholar]
- Vega, R.B.; Harrison, B.C.; Meadows, E.; Roberts, C.R.; Papst, P.J.; Olson, E.N.; McKinsey, T.A. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell Biol. 2004, 24, 8374–8385. [Google Scholar]
- Lahm, A.; Paolini, C.; Pallaoro, M.; Nardi, M.C.; Jones, P.; Neddermann, P.; Sambucini, S.; Bottomley, M.J.; Lo Surdo, P.; Carfi, A.; Koch, U.; De Francesco, R.; Steinkuhler, C.; Gallinari, P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. U S A 2007, 104, 17335–17340. [Google Scholar]
- Fischle, W.; Dequiedt, F.; Hendzel, M.J.; Guenther, M.G.; Lazar, M.A.; Voelter, W.; Verdin, E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 2002, 9, 45–57. [Google Scholar]
- Yang, X.J.; Seto, E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar]
- Yamamoto, H.; Schoonjans, K.; Auwerx, J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007, 21, 1745–1755. [Google Scholar]
- Kemper, J.K.; Xiao, Z.; Ponugoti, B.; Miao, J.; Fang, S.; Kanamaluru, D.; Tsang, S.; Wu, S.Y.; Chiang, C.M.; Veenstra, T.D. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009, 10, 392–404. [Google Scholar]
- Shimizu, T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 123–150. [Google Scholar]
- Wahli, W.; Devchand, P.R.; IJpenberg, A.; Desvergne, B. Fatty acids, eicosanoids, and hypolipidemic agents regulate gene expression through direct binding to peroxisome proliferator-activated receptors. Adv. Exp. Med. Biol. 1999, 447, 199–209. [Google Scholar]
- Biesalski, H.K. Polyphenols and inflammation: basic interactions. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 724–728. [Google Scholar]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, B.; Jones, S.A.; Price, R.R.; Watson, M.A.; McKee, D.D.; Moore, L.B.; Galardi, C.; Wilson, J.G.; Lewis, M.C.; Roth, M.E.; Maloney, P.R.; Willson, T.M.; Kliewer, S.A. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 2000, 6, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.T.; Makishima, M.; Repa, J.J.; Schoonjans, K.; Kerr, T.A.; Auwerx, J.; Mangelsdorf, D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 2000, 6, 507–515. [Google Scholar]
- Lee, Y.K.; Schmidt, D.R.; Cummins, C.L.; Choi, M.; Peng, L.; Zhang, Y.; Goodwin, B.; Hammer, R.E.; Mangelsdorf, D.J.; Kliewer, S.A. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol. Endocrinol. 2008, 22, 1345–1356. [Google Scholar]
- Kerr, T.A.; Saeki, S.; Schneider, M.; Schaefer, K.; Berdy, S.; Redder, T.; Shan, B.; Russell, D.W.; Schwarz, M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2002, 2, 713–720. [Google Scholar]
- Wang, L.; Lee, Y.K.; Bundman, D.; Han, Y.; Thevananther, S.; Kim, C.S.; Chua, S.S.; Wei, P.; Heyman, R.A.; Karin, M.; Moore, D.D. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2002, 2, 721–731. [Google Scholar]
- De Fabiani, E.; Mitro, N.; Anzulovich, A.C.; Pinelli, A.; Galli, G.; Crestani, M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J. Biol. Chem. 2001, 276, 30708–30716. [Google Scholar]
- De Fabiani, E.; Mitro, N.; Gilardi, F.; Caruso, D.; Galli, G.; Crestani, M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J. Biol. Chem. 2003, 278, 39124–39132. [Google Scholar]
- Finkel, T.; Deng, C.X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar]
- Rodgers, J.T.; Puigserver, P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U S A 2007, 104, 12861–12866. [Google Scholar]
- Scarpulla, R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar]
- Fang, S.; Miao, J.; Xiang, L.; Ponugoti, B.; Treuter, E.; Kemper, J.K. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell Biol. 2007, 27, 1407–1424. [Google Scholar]
- Kemper, J.K.; Kim, H.; Miao, J.; Bhalla, S.; Bae, Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell Biol. 2004, 24, 7707–7719. [Google Scholar]
- Boulias, K.; Talianidis, I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004, 32, 6096–6103. [Google Scholar]
- Miao, J.; Fang, S.; Lee, J.; Comstock, C.; Knudsen, K.E.; Kemper, J.K. Functional specificities of brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell Biol. 2009, 29, 6170–6181. [Google Scholar]
- Mitro, N.; Godio, C.; De Fabiani, E.; Scotti, E.; Galmozzi, A.; Gilardi, F.; Caruso, D.; Vigil Chacon, A.B.; Crestani, M. Insights in the regulation of cholesterol 7alpha-hydroxylase gene reveal a target for modulating bile acid synthesis. Hepatology 2007, 46, 885–897. [Google Scholar]
- Ananthanarayanan, M.; Li, S.; Balasubramaniyan, N.; Suchy, F.J.; Walsh, M.J. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J. Biol. Chem. 2004, 279, 54348–54357. [Google Scholar]
- Rizzo, G.; Renga, B.; Antonelli, E.; Passeri, D.; Pellicciari, R.; Fiorucci, S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol. Pharmacol. 2005, 68, 551–558. [Google Scholar]
- Miremadi, A.; Oestergaard, M.Z.; Pharoah, P.D.; Caldas, C. Cancer genetics of epigenetic genes. Hum. Mol. Genet. 2007, 16, R28–R49. [Google Scholar]
- Marks, P.; Rifkind, R.A.; Richon, V.M.; Breslow, R.; Miller, T.; Kelly, W.K. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 2001, 1, 194–202. [Google Scholar]
- Hadnagy, A.; Beaulieu, R.; Balicki, D. Histone tail modifications and noncanonical functions of histones: perspectives in cancer epigenetics. Mol. Cancer Ther. 2008, 7, 740–748. [Google Scholar]
- Gray, S.G.; De Meyts, P. Role of histone and transcription factor acetylation in diabetes pathogenesis. Diabetes Metab. Res. Rev. 2005, 21, 416–433. [Google Scholar]
- Potthoff, M.J.; Wu, H.; Arnold, M.A.; Shelton, J.M.; Backs, J.; McAnally, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 2007, 117, 2459–2467. [Google Scholar]
- McGee, S.L.; van Denderen, B.J.; Howlett, K.F.; Mollica, J.; Schertzer, J.D.; Kemp, B.E.; Hargreaves, M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes 2008, 57, 860–867. [Google Scholar]
- Czubryt, M.P.; McAnally, J.; Fishman, G.I.; Olson, E.N. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha ) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA 2003, 100, 1711–1716. [Google Scholar]
- Zhang, Y.; Talalay, P. Anticarcinogenic activities of organic isothiocyanates: chemistry and mechanisms. Cancer Res. 1994, 54, 1976s–1981s. [Google Scholar]
- Hecht, S.S. Chemoprevention by isothiocyanates. J. Cell Biochem. Suppl. 1995, 22, 195–209. [Google Scholar]
- Nian, H.; Delage, B.; Ho, E.; Dashwood, R.H. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ. Mol. Mutagen. 2009, 50, 213–221. [Google Scholar]
- Myzak, M.C.; Karplus, P.A.; Chung, F.L.; Dashwood, R.H. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004, 64, 5767–5774. [Google Scholar]
- Myzak, M.C.; Dashwood, W.M.; Orner, G.A.; Ho, E.; Dashwood, R.H. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006, 20, 506–508. [Google Scholar]
- Myzak, M.C.; Hardin, K.; Wang, R.; Dashwood, R.H.; Ho, E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 2006, 27, 811–819. [Google Scholar]
- Nian, H.; Delage, B.; Pinto, J.T.; Dashwood, R.H. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis 2008, 29, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Englyst, H.N. Fermentation in the human large intestine and the available substrates. Am. J. Clin. Nutr. 1987, 45, 1243–1255. [Google Scholar]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; Scherer, B.; Sinclair, D.A. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar]
- Lagouge, M.; Argmann, C.; Gerhart-Hines, Z.; Meziane, H.; Lerin, C.; Daussin, F.; Messadeq, N.; Milne, J.; Lambert, P.; Elliott, P.; Geny, B.; Laakso, M.; Puigserver, P.; Auwerx, J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006, 127, 1109–1122. [Google Scholar]
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; Pistell, P.J.; Poosala, S.; Becker, K.G.; Boss, O.; Gwinn, D.; Wang, M.; Ramaswamy, S.; Fishbein, K.W.; Spencer, R.G.; Lakatta, E.G.; Le Couteur, D.; Shaw, R.J.; Navas, P.; Puigserver, P.; Ingram, D.K.; de Cabo, R.; Sinclair, D.A. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006, 444, 337–342. [Google Scholar]
- Canto, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar]
- Borra, M.T.; Smith, B.C.; Denu, J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005, 280, 17187–17195. [Google Scholar]
- Kaeberlein, M.; McDonagh, T.; Heltweg, B.; Hixon, J.; Westman, E.A.; Caldwell, S.D.; Napper, A.; Curtis, R.; DiStefano, P.S.; Fields, S.; Bedalov, A.; Kennedy, B.K. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005, 280, 17038–17045. [Google Scholar]
- Zunino, S. Type 2 diabetes and glycemic response to grapes or grape products. J. Nutr. 2009, 139, 1794S–1800S. [Google Scholar]
- Rasbach, K.A.; Schnellmann, R.G. Isoflavones promote mitochondrial biogenesis. J. Pharmacol. Exp. Ther. 2008, 325, 536–543. [Google Scholar]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
De Fabiani, E.; Mitro, N.; Gilardi, F.; Galmozzi, A.; Caruso, D.; Crestani, M. When Food Meets Man: the Contribution of Epigenetics to Health. Nutrients 2010, 2, 551-571. https://doi.org/10.3390/nu2050551
De Fabiani E, Mitro N, Gilardi F, Galmozzi A, Caruso D, Crestani M. When Food Meets Man: the Contribution of Epigenetics to Health. Nutrients. 2010; 2(5):551-571. https://doi.org/10.3390/nu2050551
Chicago/Turabian StyleDe Fabiani, Emma, Nico Mitro, Federica Gilardi, Andrea Galmozzi, Donatella Caruso, and Maurizio Crestani. 2010. "When Food Meets Man: the Contribution of Epigenetics to Health" Nutrients 2, no. 5: 551-571. https://doi.org/10.3390/nu2050551
APA StyleDe Fabiani, E., Mitro, N., Gilardi, F., Galmozzi, A., Caruso, D., & Crestani, M. (2010). When Food Meets Man: the Contribution of Epigenetics to Health. Nutrients, 2(5), 551-571. https://doi.org/10.3390/nu2050551