Pilot Dietary Intervention with Heat-Stabilized Rice Bran Modulates Stool Microbiota and Metabolites in Healthy Adults
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pilot Trial Design and Participation
2.2. Nutritional Composition of SRB
2.2.1. Heat Stabilization of Rice Bran
2.2.2. Composition of SRB and Control Intervention Meals and Snacks
2.3. Pyrosequencing of the Bacterial Community
2.3.1. DNA Extraction, Amplification and Sequencing
2.3.2. Analysis of Microbiota
2.4. Metabolite Profiling
2.4.1. Metabolite Extraction and Detection by Gas Chromatography-Mass Spectrometry
2.4.2. Short Chain Fatty Acid Determination
2.5. Statistical Analysis and Data Visualization
2.5.1. Microbiota Analyses
2.5.2. Metabolome Analyses
3. Results
3.1. Increased SRB Effects on Caloric and Macronutrient Intakes
| Characteristic | Control (n = 3) | Rice Bran (n = 4) |
|---|---|---|
| Age (years) a | 42.3 ± 21.7 | 42.8 ± 15.6 |
| Sex | ||
| Males (%) | 2 (67%) | 0 (0%) |
| Females (%) | 1 (33%) | 4 (100%) |
| BMI (kg/m2) a | 28.9 ± 6.9 | 22 ± 1.7 |
| Total cholesterol a (mg/dL) | 187 ± 57.2 | 197 ± 54.6 |
| LDL a (mg/dL) | 118 ± 50.3 | 127 ± 40.9 |
| HDL a (mg/dL) | 44 ± 12.6 | 54.3 ± 17.6 |
| Triglycerides a (mg/dL) | 125.7 ± 80.0 | 80 ± 35.0 |
| Fruit intake (X servings/day) b | ||
| 0 ≤ X ≤ 2 | 2 | 3 |
| X > 2 | 1 | 1 |
| Vegetable intake (X servings/day) b | ||
| 0 ≤ X ≤ 2 | 1 | 1 |
| X > 2 | 2 | 3 |
| Grain intake (X servings/day) b | ||
| 0 ≤ X ≤ 4 | 2 | 4 |
| X > 4 | 1 | 0 |

| Dietary Intake | Control | Rice Bran | ||
|---|---|---|---|---|
| Week 2 | Week 4 | Week 2 | Week 4 | |
| Calories (kcal) | 2015.3 ± 325.0 (2186.4) | 2047.8 ± 265.6 (2099.1) | 2052.9 ± 410.3 (1940.6) | 1925.3 ± 335.5 (1791.4) |
| Protein (g) | 81.7 ± 13.7 (80.1) | 77.6 ± 17.9 (77.6) b | 86.3 ± 14.0 (85.3) | 68.9 ± 9.9 (71.1) b |
| Carbohydrates (g) | 264.6 ± 54.0 (290.8) | 267.6 ± 53.0 (277.3) a | 253.0 ± 46.4 (243.9) | 255.6 ± 58.3 (241.4) a |
| Fat (g) | 67.1 ± 13.9 (72.4) | 74.6 ± 12.3 (81.0) b | 79.8 ± 16.6 (74.2) | 75.4 ± 13.3 (74.3) a |
| Fiber (g) | 24.2 ± 3.0 (22.8) a | 23.5 ± 8.0 (19.4) b | 36.0 ± 7.5 (35.7) a | 32.4 ± 5.6 (31.9) b |
3.2. Microbiome Changes with Consumption of SRB


| Closest Hit in Database | 2 weeks | q-Value | 4 weeks | q-Value |
|---|---|---|---|---|
| Methanobrevibacter smithii | 1201.00% | <0.001 | 210.73% | <0.001 |
| Paraprevotella clara | 352.87% | <0.001 | 156.71% | <0.001 |
| Ruminococcus flavefaciens | 128.49% | <0.001 | 79.02% | <0.001 |
| Dialister succinatiphilus | 86.59% | <0.001 | 57.47% | <0.001 |
| Bifidobacterium sp. | 2.79% | 1.000 | 50.29% | 0.003 |
| Clostridium glycolicum (Clostridium cluster XI) | 0.00% | 1.000 | 40.71% | 0.042 |
| Barnesiella intestinihominis | 277.35% | <0.001 | 66.31% | 0.050 |
| Anaerostipes caccae | 90.09% | <0.001 | 69.63% | 0.483 |
| Ruminococcus bromii | 66.77% | <0.001 | 29.47% | 1.000 |
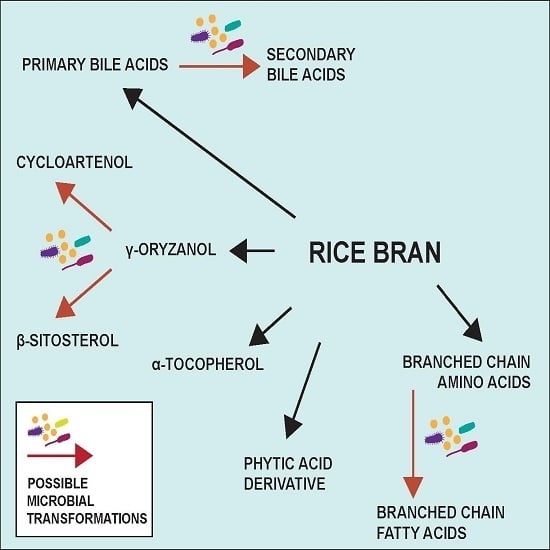
3.3. Metabolome Changes with Increased SRB

| Stool Metabolites | % change at 4 weeks | KEGG pathway |
|---|---|---|
| Amino acids and nucleosides | ||
| Inosine | 3.72% | Purine metabolism |
| Uridine | 3.22% | Pyrimidine metabolism |
| Glutamic acid * | 1.82% | Purine and pyrimidine metabolism |
| Glutaric acid | 1.73% | Lysine degradation |
| Glycine * | −1.56% | Purine metabolism |
| Leucine * | −3.75% | Amino acid metabolism |
| Cholesterol and bile acids | ||
| Cholest-8(14)-en-3-one | 6.78% | N/A |
| Deoxycholic acid | 2.69% | Secondary bile acid biosynthesis |
| Cholest5-en-3-ol-propionate | 2.12% | N/A |
| Lithocholic acid | 1.07% | Secondary bile acid biosynthesis |
| Cholesterol | 0.51% | Steroid biosynthesis |
| Phytochemicals and phenolics | ||
| Indole-2-carboxylic acid * | 11.65% | N/A |
| Hydrocinnamic acid | 4.31% | Phenylalanine metabolism |
| Alpha-tocopherol * | 2.46% | Vitamin digestion and absorption |
| Benzoic acid | 2.39% | Phenylalanine metabolism |
| Cycloartenol * | 1.90% | Steroid biosynthesis |
| Pantothenic acid * | 1.90% | Vitamin digestion and absorption |
| Phenylacetic acid | 1.49% | Phenylalanine metabolism |
| Beta-sitosterol * | 0.11% | Steroid biosynthesis |
| Lipids | ||
| Myristic acid * | 7.32% | Fatty acid biosynthesis |
| Caprylic acid | 3.84% | Fatty acid biosynthesis |
| Lauric acid | 3.03% | Fatty acid biosynthesis |
| Palmitic acid * | 2.20% | Fatty acid biosynthesis |
| Stearic acid * | 1.12% | Fatty acid biosynthesis |
| Azelaic acid | 0.56% | N/A |
| Glycerol | 0.55% | Galactose metabolism |
| Oleic acid * | 0.15% | Fatty acid biosynthesis |
| Sebacic acid | −0.33% | N/A |
| 2-Hexenedioic acid | −0.32% | N/A |
| Pentadecanoic acid | −1.90% | N/A |
| Putative microbial metabolites | ||
| Indole-2-carboxylic acid * | 11.65% | N/A |
| Hydrocinnamic acid a | 4.31% | Phenylalanine metabolism |
| Inositol monophosphate a | 3.90% | Inositol phosphate metabolism |
| Phosphoric acid a | 3.61% | Peptidoglycan synthesis |
| Deoxycholic acid | 2.69% | Secondary bile acid biosynthesis |
| Putative microbial metabolites | ||
| Benzoic acid a | 2.39% | Phenylalanine metabolism |
| Cycloartenol a | 1.90% | Steroid biosynthesis |
| Phenylacetic acid a | 1.49% | Phenylalanine metabolism |
| Stearic acid a | 1.12% | Fatty acid biosynthesis |
| Lithocholic acid | 1.07% | Secondary bile acid biosynthesis |
| Beta-sitosterol a | 0.11% | Steroid biosynthesis |
| Sugars b | ||
| Maltose | −0.10% | Carbohydrate digestion |
| Ribose | −3.56% | Carbohydrate digestion |
| Glucose | −3.63% | Carbohydrate digestion |
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cicero, A.; Derosa, G. Rice bran and its main components: Potential role in the management of coronary risk factors. Curr. Top. Nutraceutical Res. 2005, 3, 29–46. [Google Scholar]
- Cheng, H.-H.; Huang, H.-Y.; Chen, Y.-Y.; Huang, C.-L.; Chang, C.-J.; Chen, H.-L.; Lai, M.-H. Ameliorative effects of stabilized rice bran on type 2 diabetes patients. Ann. Nutr. Metabol. 2009, 56, 45–51. [Google Scholar] [CrossRef]
- De Munter, J.S.; Hu, F.B.; Spiegelman, D.; Franz, M.; van Dam, R.M. Whole grain, bran, and germ intake and risk of type 2 diabetes: A prospective cohort study and systematic review. PLoS Med. 2007, 4, e261. [Google Scholar] [CrossRef]
- Jariwalla, R. Rice-bran products: Phytonutrients with potential applications in preventive and clinical medicine. Drugs Exp. Clin. Res. 2000, 27, 17–26. [Google Scholar]
- Kim, T.H.; Kim, E.K.; Lee, M.-S.; Lee, H.-K.; Hwang, W.S.; Choe, S.J.; Kim, T.-Y.; Han, S.J.; Kim, H.J.; Kim, D.J. Intake of brown rice lees reduces waist circumference and improves metabolic parameters in type 2 diabetes. Nutr. Res. 2011, 31, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.J.; Ollila, C.A.; Kumar, A.; Borresen, E.C.; Raina, K.; Agarwal, R.; Ryan, E.P. Chemopreventive properties of dietary rice bran: Current status and future prospects. Adv. Nutr. 2012, 3, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Phutthaphadoong, S.; Yamada, Y.; Hirata, A.; Tomita, H.; Hara, A.; Limtrakul, P.; Iwasaki, T.; Kobayashi, H.; Mori, H. Chemopreventive effect of fermented brown rice and rice bran (FBRA) on the inflammation-related colorectal carcinogenesis in ApcMin/+ mice. Oncol. Rep. 2010, 23, 53–59. [Google Scholar] [PubMed]
- Verschoyle, R.; Greaves, P.; Cai, H.; Edwards, R.; Steward, W.; Gescher, A. Evaluation of the cancer chemopreventive efficacy of rice bran in genetic mouse models of breast, prostate and intestinal carcinogenesis. Br. J. Cancer 2007, 96, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Kahlon, T.S. Rice Bran: Production, Composition, Functionality and Food Applications, Physiological Benefits; Taylor and Francis Group, LLC: Boca Raton, FL, USA, 2009. [Google Scholar]
- Prasad, M.N. Health benefits of rice bran—A review. J. Nutr. Food Sci. 2011, 1, 108. [Google Scholar] [CrossRef]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A.; et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013, 7, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Ogué-Bon, E.; Khoo, C.; McCartney, A.L.; Gibson, G.R.; Rastall, R.A. In vitro effects of synbiotic fermentation on the canine faecal microbiota. FEMS Microbiol. Ecol. 2010, 73, 587–600. [Google Scholar] [PubMed]
- Komiyama, Y.; Andoh, A.; Fujiwara, D.; Ohmae, H.; Araki, Y.; Fujiyama, Y.; Mitsuyama, K.; Kanauchi, O. New prebiotics from rice bran ameliorate inflammation in murine colitis models through the modulation of intestinal homeostasis and the mucosal immune system. Scand. J. Gastroenterol. 2011, 46, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.V.; Ockhuizen, T. New insights into the impact of the intestinal microbiota on health and disease: A symposium report. Br. J. Nutr. 2012, 107, S1–S13. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J. The impact of nutrition on the human microbiome. Nutr. Rev. 2012, 70, S10–S13. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Arora, T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-promoting effects of microbial dysbiosis. Curr. Oncol. Rep. 2014, 16, 1–9. [Google Scholar] [CrossRef]
- Suzuki, T.; Yoshida, S.; Hara, H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br. J. Nutr. 2008, 100, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Vipperla, K.; O’Keefe, S.J. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr. Clin. Pract. 2012, 27, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Waldecker, M.; Kautenburger, T.; Daumann, H.; Busch, C.; Schrenk, D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008, 19, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One 2013, 8, e70803. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Sheneman, L.; Evans, J.; Foster, J.A. Clearcut: A fast implementation of relaxed neighbor joining. Bioinformatics 2006, 22, 2823–2824. [Google Scholar] [CrossRef] [PubMed]
- European Nucleotide Archive. Available online: http://www.ebi.ac.uk/ena/data/view/PRJEB8075 (accessed on 12 February 2015).
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- NIST/EPA/NIH Mass Spectral Database, NIST 11. Available online: http://chemdata.nist.gov/ (accessed on 25 August 2013).
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. [email protected]: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Davies, T. The new automated mass spectrometry deconvolution and identification system (AMDIS). Spectrosc. Eur. 1998, 10, 24–27. [Google Scholar]
- The R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://web.mit.edu/r_v3.0.1/fullrefman.pdf (accessed on 25 August 2013).
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comp. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Agriculture, A.R.S. Nutrient Data Laboratory Home Page. USDA National Nutrient Database for Standard Reference, Release 26. Available online: http://www.ars.usda.gov/ba/bhnrc/ndl (accessed on 5 February 2013).
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [PubMed]
- Vernocchi, P.; Vannini, L.; Gottardi, D.; Del Chierico, F.; Serrazanetti, D.I.; Ndagijimana, M.; Guerzoni, M.E. Integration of datasets from different analytical techniques to assess the impact of nutrition on human metabolome. Front. Cell. Infect. Microbiol. 2012, 2, 156. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Forster, G.M.; Raina, K.; Kumar, A.; Kumar, S.; Agarwal, R.; Chen, M.-H.; Bauer, J.E.; McClung, A.M.; Ryan, E.P. Rice varietal differences in bioactive bran components for inhibition of colorectal cancer cell growth. Food Chem. 2013, 141, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, H.; Ikata, K.; Arimochi, H.; Iwasaki, T.; Ohnishi, Y.; Kuwahara, T.; Kataoka, K. Effects of fermented brown rice on the intestinal environments in healthy adult. J. Investig. Med. 2011, 58, 235–245. [Google Scholar] [CrossRef]
- Yang, J.; Keshavarzian, A.; Rose, D.J. Impact of dietary fiber fermentation from cereal grains on metabolite production by the fecal microbiota from normal weight and obese individuals. J. Med. Food 2013, 16, 862–867. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Mariotti, F.; Huneau, J.; Tome, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar]
- Boudry, G.; Jamin, A.; Chatelais, L.; Gras-Le Guen, C.; Michel, C.; Le Huërou-Luron, I. Dietary protein excess during neonatal life alters colonic Microbiota and mucosal response to inflammatory mediators later in life in female pigs. J. Nutr. 2013, 143, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Allison, M.; Bryant, M.; Katz, I.; Keeney, M. Metabolic function of branched-chain volatile fatty acids, growth factors for ruminococci II. Biosynthesis of higher branched-chain fatty acids and aldehydes. J. Bacteriol. 1962, 83, 1084–1093. [Google Scholar] [PubMed]
- Ran-Ressler, R.R.; Khailova, L.; Arganbright, K.M.; Adkins-Rieck, C.K.; Jouni, Z.E.; Koren, O.; Ley, R.E.; Brenna, J.T.; Dvorak, B. Branched chain fatty acids reduce the incidence of necrotizing enterocolitis and alter gastrointestinal microbial ecology in a neonatal rat model. PloS One 2011, 6, e29032. [Google Scholar] [CrossRef]
- Hu, G.; Yu, W. Binding of cholesterol and bile acid to hemicelluloses from rice bran. Int. J. Food Sci. Nutr. 2013, 64, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Gestel, G.; Besancon, P.; Rouanet, J.-M. Comparative evaluation of the effects of two different forms of dietary fibre (rice bran vs. wheat bran) on rat colonic mucosa and faecal microflora. Ann. Nutr. Metabol. 1994, 38, 249–256. [Google Scholar] [CrossRef]
- Robson, J. Lipid modification: Cardiovascular risk assessment and the modification of blood lipids for the primary and secondary prevention of cardiovascular disease. Heart 2008, 94, 1331–1332. [Google Scholar] [CrossRef] [PubMed]
- Rukmini, C.; Raghuram, T.C. Nutritional and biochemical aspects of the hypolipidemic action of rice bran oil: A review. J. Am. Coll. Nutr. 1991, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Yasukawa, K.; Yamaura, M.; Ukiya, M.; Kimura, Y.; Shimizu, N.; Arai, K. Triterpene alcohol and sterol ferulates from rice bran and their anti-inflammatory effects. J. Agric. Food Chem. 2000, 48, 2313–2319. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, K.; Akihisa, T.; Kimura, Y.; Tamura, T.; Takido, M. Inhibitory effect of cycloartenol ferulate, a component of rice bran, on tumor promotion in two-stage carcinogenesis in mouse skin. Biol. Pharm. Bull. 1998, 21, 1072–1076. [Google Scholar] [CrossRef] [PubMed]
- Baskar, A.A.; Al Numair, K.S.; Gabriel Paulraj, M.; Alsaif, M.A.; Muamar, M.A.; Ignacimuthu, S. Beta-sitosterol prevents lipid peroxidation and improves antioxidant status and histoarchitecture in rats with 1, 2-dimethylhydrazine-induced colon cancer. J. Med. Food 2012, 15, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.C.; Huss, U.; Jenner, A.; Halliwell, B.; Bohlin, L.; Rafter, J.J. Human fecal water inhibits COX-2 in colonic HT-29 cells: Role of phenolic compounds. J. Nutr. 2005, 135, 2343–2349. [Google Scholar] [PubMed]
- Wang, T.; Hicks, K.B.; Moreau, R. Antioxidant activity of phytosterols, oryzanol, and other phytosterol conjugates. J. Am. Oil Chem. Soc. 2002, 79, 1201–1206. [Google Scholar] [CrossRef]
- Markiewicz, L.H.; Honke, J.; Haros, M.; Świątecka, D.; Wróblewska, B. Diet shapes the ability of human intestinal microbiota to degrade phytate—In vitro studies. J. Appl. Microbiol. 2013, 115, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiology Reviews 2010, 34, 426–444. [Google Scholar] [PubMed]
- Braune, A.; Bunzel, M.; Yonekura, R.; Blaut, M. Conversion of dehydrodiferulic acids by human intestinal microbiota. J. Agric. Food Chem. 2009, 57, 3356–3362. [Google Scholar] [CrossRef] [PubMed]
- Vetrani, C.; Rivellese, A.A.; Annuzzi, G.; Mattila, I.; Meudec, E.; Hyötyläinen, T.; Orešič, M.; Aura, A.-M. Phenolic metabolites as compliance biomarker for polyphenol intake in a randomized controlled human intervention. Food Res. Int. 2014, 63, 233–238. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheflin, A.M.; Borresen, E.C.; Wdowik, M.J.; Rao, S.; Brown, R.J.; Heuberger, A.L.; Broeckling, C.D.; Weir, T.L.; Ryan, E.P. Pilot Dietary Intervention with Heat-Stabilized Rice Bran Modulates Stool Microbiota and Metabolites in Healthy Adults. Nutrients 2015, 7, 1282-1300. https://doi.org/10.3390/nu7021282
Sheflin AM, Borresen EC, Wdowik MJ, Rao S, Brown RJ, Heuberger AL, Broeckling CD, Weir TL, Ryan EP. Pilot Dietary Intervention with Heat-Stabilized Rice Bran Modulates Stool Microbiota and Metabolites in Healthy Adults. Nutrients. 2015; 7(2):1282-1300. https://doi.org/10.3390/nu7021282
Chicago/Turabian StyleSheflin, Amy M., Erica C. Borresen, Melissa J. Wdowik, Sangeeta Rao, Regina J. Brown, Adam L. Heuberger, Corey D. Broeckling, Tiffany L. Weir, and Elizabeth P. Ryan. 2015. "Pilot Dietary Intervention with Heat-Stabilized Rice Bran Modulates Stool Microbiota and Metabolites in Healthy Adults" Nutrients 7, no. 2: 1282-1300. https://doi.org/10.3390/nu7021282
APA StyleSheflin, A. M., Borresen, E. C., Wdowik, M. J., Rao, S., Brown, R. J., Heuberger, A. L., Broeckling, C. D., Weir, T. L., & Ryan, E. P. (2015). Pilot Dietary Intervention with Heat-Stabilized Rice Bran Modulates Stool Microbiota and Metabolites in Healthy Adults. Nutrients, 7(2), 1282-1300. https://doi.org/10.3390/nu7021282