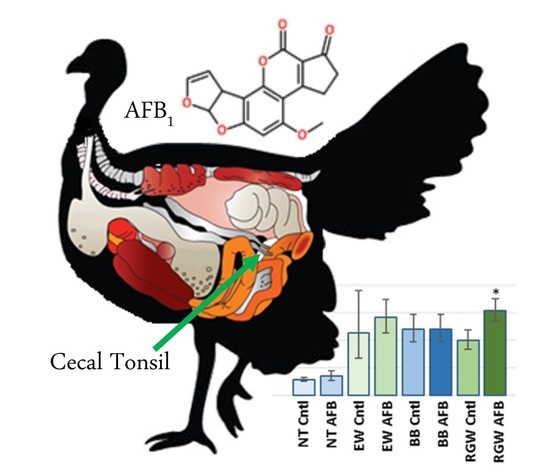
Differential Transcriptome Responses to Aflatoxin B1 in the Cecal Tonsil of Susceptible and Resistant Turkeys
Abstract
:1. Introduction
2. Results
2.1. Differential Gene Expression
2.1.1. AFB1 Treatment Effects
Unique Transcriptome Responses
2.2. Wild versus Domesticated Turkey
2.2.1. Control Birds
2.2.2. AFB1 Treatment
3. Discussion
4. Materials and Methods
4.1. RNA Isolation and Sequencing
4.2. Quantitative Real-Time PCR
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFB | aflatoxin B1 |
| AFBO | exo-AFB1-8,9-epoxide |
| BB | Broad Breasted White |
| Ct | threshold cycle |
| CYP | cytochrome P450 |
| DE | differentially expressed |
| DEG | differentially expressed gene |
| DT | domesticated turkey |
| EW | Eastern wild turkey (Meleagris gallopavo silvestris) |
| FC | fold change |
| FDR | false discovery rate |
| GO | gene ontology |
| GST | glutathione S-transferase |
| IPA | Ingenuity Pathway Analysis |
| ncRNA | non-coding RNA |
| PCA | principal component analysis |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| RGW | Rio Grande wild turkey (Meleagris gallopavo intermedia) |
References
- CAST. Mycotoxins: Risks in Plant, Animal and Human Systems; No. 139; Council for Agricultural Science and Technology: Ames, IA, USA, 2003. [Google Scholar]
- Blount, W.P. Turkey “X” disease. Turkeys 1961, 9, 52, 55–58, 61–71, 77. [Google Scholar]
- Rawal, S.; Kim, J.E.; Coulombe, R., Jr. Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Res. Vet. Sci. 2010, 89, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Coulombe, R.A.; Reed, K.M. Aflatoxicosis: Lessons from toxicity and responses to aflatoxin B1 in poultry. Agriculture 2015, 5, 742–777. [Google Scholar] [CrossRef]
- Qureshi, M.A.; Brake, J.; Hamilton, P.B.; Hagler, W.M., Jr.; Nesheim, S. Dietary exposure of broiler breeders to aflatoxin results in immune dysfunction in progeny chicks. Poult. Sci. 1998, 77, 812–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qureshi, M.A.; Heggen, C.L.; Hussain, I. Avian macrophage: Effector functions in health and disease. Dev. Comp. Immunol. 2000, 24, 103–119. [Google Scholar] [CrossRef]
- Williams, J.G.; Deschl, U.; Williams, G.M. DNA damage in fetal liver cells of turkey and chicken eggs dosed with aflatoxin B1. Arch. Toxicol. 2011, 85, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Quist, C.F.; Bounous, D.I.; Kilburn, J.V.; Nettles, V.F.; Wyatt, R.D. The effect of dietary aflatoxin on wild turkey poults. J. Wildl. Dis. 2000, 36, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Rawal, S.; Coulombe, R.A., Jr. Metabolism of aflatoxin B1 in turkey liver microsomes: The relative roles of cytochromes P450 1A5 and 3A37. Toxicol. Appl. Pharmacol. 2011, 254, 349–354. [Google Scholar] [CrossRef]
- Bunderson, B.R.; Kim, J.E.; Croasdell, A.; Mendoza, K.M.; Reed, K.M.; Coulombe, R.A., Jr. Heterologous expression and functional characterization of avian mu-class glutathione S-transferases. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2013, 158, 109–116. [Google Scholar] [CrossRef]
- Kim, J.E.; Bunderson, B.R.; Croasdell, A.; Coulombe, R.A., Jr. Functional characterization of alpha-class glutathione S-transferases from the turkey (Meleagris gallopavo). Toxicol. Sci. 2011, 124, 45–53. [Google Scholar] [CrossRef]
- Kim, J.E.; Bunderson, B.R.; Croasdell, A.; Reed, K.M.; Coulombe, R.A., Jr. Alpha-class glutathione S-transferases in wild turkeys (Meleagris gallopavo): Characterization and role in resistance to the carcinogenic mycotoxin aflatoxin B1. PLoS ONE 2013, 8, e60662. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.J.; Buckner, R.; Kelly, J.; Coulombe, R.A. Biochemical basis for the extreme sensitivity of turkeys to aflatoxin B1. Toxicol. Appl. Pharmacol. 2000, 165, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.J.; Van Vleet, T.R.; Hall, J.O.; Coulombe, R.A., Jr. Dietary butylated hydroxytoluene protects against aflatoxicosis in turkeys. Toxicol. Appl. Pharmacol. 2002, 182, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Settlage, R.E.; McMahon, K.W.; Mendoza, K.M.; Rawal, S.; El-Nemazi, H.S.; Coulombe, R.A., Jr.; Reed, K.M. Response of the hepatic transcriptome to aflatoxin B1 in domestic turkey (Meleagris gallopavo). PLoS ONE 2014, 9, e100930. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Settlage, R.E.; Mendoza, K.M.; Rawal, S.; El-Nezami, H.S.; Coulombe, R.A., Jr.; Reed, K.M. Modulation of the spleen transcriptome in domestic turkey (Meleagris gallopavo) in response to aflatoxin B1 and probiotics. Immunogenetics 2015, 67, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.M.; Mendoza, K.M.; Abrahante, J.E.; Coulombe, R.A. Comparative response of the hepatic transcriptomes of domesticated and wild turkey to aflatoxin B1. Toxins 2018, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Cardona, C.C.; Coulombe, R.A.; Reed, K.M. Hepatic transcriptome responses of domesticated and wild turkey embryos to aflatoxin B1. Toxins 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Grenier, B.; Applegate, T.J. Modulation of intestinal functions following mycotoxin ingestion: Meta-analysis of published experiments in animals. Toxins 2013, 5, 396–430. [Google Scholar] [CrossRef] [PubMed]
- Galarza-Seeber, R.; Latorre, J.D.; Bielke, L.R.; Kuttappan, V.A.; Wolfenden, A.D.; Hernandez-Velasco, X.; Merino-Guzman, R.; Vicente, J.L.; Donoghue, A.; Cross, D.; et al. Leaky gut and mycotoxins: Aflatoxin B1 does not increase gut permeability in broiler chickens. Front. Vet. Sci. 2016, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Naehrer, K.; Applegate, T.J. Interactive effects of dietary protein concentration and aflatoxin B1 on performance, nutrient digestibility and gut health in broiler chicks. Poult. Sci. 2016, 95, 1312–1325. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Denman, S.E.; Hughes, R.J.; Geier, M.S.; Crowley, T.M.; Chen, H.; Haring, V.R.; Moore, R.J. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 2012, 96, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Matsui, C.; Furuse, K.; Mimori-Kiyosue, Y.; Furuse, M.; Tsukita, S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc. Natl. Acad. Sci. USA 2003, 100, 3971–3976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grishin, A.V.; Sverdlov, V.E.; Kostina, M.B.; Modyanov, N.N. Cloning and characterization of the entire cDNA encoded by ATP1AL1--a member of the human Na,K/H,K-ATPase gene family. FEBS Lett. 1994, 349, 144–150. [Google Scholar] [CrossRef]
- Hinson, E.R.; Cresswell, P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 2009, 284, 4705–4712. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, L.; Schmidt, R.E.; Su, C.; Huang, X.; Gould, K.; Cao, G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005, 332, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Grahn, T.H.; Zhang, Y.; Lee, M.J.; Sommer, A.G.; Mostoslavsky, G.; Fried, S.K.; Greenberg, A.S.; Puri, V. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem. Biophys. Res. Commun. 2013, 432, 296–301. [Google Scholar] [CrossRef] [Green Version]
- Feild, J.A.; Zhang, L.; Brun, K.A.; Brooks, D.P.; Edwards, R.M. Cloning and functional characterization of a sodium-dependent phosphate transporter expressed in human lung and small intestine. Biochem. Biophys. Res. Commun. 1999, 258, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Brady, J.R.; Chan, B.; Lee, W.P.; Hsu, B.; Harless, S.; Cancro, M.; Grewal, I.S.; Dixit, V.M. Identification of a novel receptor for B lymphocyte stimulator that is mutated in a mouse strain with severe B cell deficiency. Curr. Biol. 2001, 11, 1547–1552. [Google Scholar] [CrossRef] [Green Version]
- Spreckley, E.; Murphy, K.G. The L-cell in nutritional sensing and the regulation of appetite. Front. Nutr. 2015, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Karniski, L.P.; Aronson, P.S. Chloride/formate exchange with formic acid recycling: A mechanism of active chloride transport across epithelial membranes. Proc. Natl. Acad. Sci. USA 1985, 82, 6362–6365. [Google Scholar] [CrossRef]
- Racioppi, L.; Means, A.R. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveler. Trends Immunol. 2008, 29, 600–607. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007, 81, 11246–11255. [Google Scholar] [CrossRef]
- Mulero, J.J.; Boyle, B.J.; Bradley, S.; Bright, J.M.; Nelken, S.T.; Ho, T.T.; Mize, N.K.; Childs, J.D.; Ballinger, D.G.; Ford, J.E.; et al. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP). Immunogenetics 2002, 54, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria-host communication: The language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956. [Google Scholar] [CrossRef] [Green Version]
- Prabhu, A.V.; Luu, W.; Sharpe, L.J.; Brown, A.J. Phosphorylation regulates activity of 7-dehydrocholesterol reductase (DHCR7), a terminal enzyme of cholesterol synthesis. J. Steroid Biochem. Mol. Biol. 2017, 165, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.J.; Skurat, A.V. Self-glucosylating initiator proteins and their role in glycogen biosynthesis. Prog. Nucl. Acid Res. Mol. Biol. 1997, 57, 289–316. [Google Scholar]
- Braun, T.; Voland, P.; Kunz, L.; Prinz, C.; Gratzl, M. Enterochromaffin cells of the human gut: Sensors for spices and odorants. Gastroenterology 2007, 132, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Sternini, C.; Anselmi, L.; Rozengurt, E. Enteroendocrine cells: A site of ‘taste’ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008, 15, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef]
- Coulombe, R.A., Jr. Biological action of mycotoxins. J. Dairy Sci. 1993, 76, 880–891. [Google Scholar] [CrossRef]
- Kumagai, S. Intestinal absorption and excretion of aflatoxin in rats. Toxicol. Appl. Pharmacol. 1989, 97, 88–97. [Google Scholar] [CrossRef]
- Larsson, P.; Tjälve, H. Intranasal instillation of aflatoxin B1 in rats: Bioactivation in the nasal mucosa and neuronal transport to the olfactory bulb. Toxicol. Sci. 2000, 55, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.; Wu, Q.K.; El-Nezami, H.; Juvonen, R.O.; Mykkänen, H.; Turner, P.C. Lactobacillus rhamnosus strain GG reduces aflatoxin B1 transport, metabolism and toxicity in Caco-2 cells. Appl. Environ. Microbiol. 2007, 73, 3958–3964. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Ares, I.; Ramos, E.; Castellano, V.; Martínez, M.; Martínez-Larrañaga, M.R.; Anadón, A.; Martínez, M.A. Mycotoxins modify the barrier function of Caco-2 cells through differential gene expression of specific claudin isoforms: Protective effect of illite mineral clay. Toxicology 2016, 353–354, 21–33. [Google Scholar] [CrossRef]
- Caloni, F.; Cortinovis, C.; Pizzo, F.; De Angelis, I. Transport of aflatoxin M1 in human intestinal caco-2/TC7 cells. Front. Pharmacol. 2012, 3, 111. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.Y.; Voellinger, J.L.; Van Ness, K.P.; Chapron, B.; Shaffer, R.M.; Neumann, T.; White, C.C.; Kavanagh, T.J.; Kelly, E.J.; Eaton, D.L. Characterization of rat or human hepatocytes cultured in microphysiological systems (MPS) to identify hepatotoxicity. Toxicol. In Vitro 2017, 40, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M.; Madsen, K.; Spiller, R.; Van Meerveld, B.G.; Verne, G.N. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol. Motil. 2012, 24, 503–512. [Google Scholar] [CrossRef] [Green Version]
- Akbari, P.; Braber, S.; Varasteh, S.; Alizadeh, A.; Garssen, J.; Fink-Gremmels, J. The intestinal barrier as an emerging target in the toxicological assessment of mycotoxins. Arch. Toxicol. 2017, 91, 1007–1029. [Google Scholar] [CrossRef]
- Liew, W.P.; Mohd-Redzwan, S. Mycotoxin: Its impact on gut health and microbiota. Front. Cell Infect. Microbiol. 2018, 8, 60. [Google Scholar] [CrossRef]
- Robert, H.; Payros, D.; Pinton, P.; Théodorou, V.; Mercier-Bonin, M.; Oswald, I.P. Impact of mycotoxins on the intestine: Are mucus and microbiota new targets? J. Toxicol. Environ. Health B Crit. Rev. 2017, 20, 249–275. [Google Scholar]
- Ishikawa, A.T.; Weese, J.S.; Bracarense, A.P.F.R.L.; Alfieri, A.A.; Oliveira, G.G.; Kawamura, O.; Hirooka, E.Y.; Itano, E.N.; Costa, M.C. Single aflatoxin B1 exposure induces changes in gut microbiota community in C57Bl/6 mice. World Mycotoxin J. 2017, 10, 249–254. [Google Scholar] [CrossRef]
- Wang, J.; Tang, L.; Glenn, T.C.; Wang, J.S. Aflatoxin B1 induced compositional changes in gut microbial communities of male F344 rats. Toxicol. Sci. 2016, 150, 54–63. [Google Scholar] [CrossRef] [PubMed]
- El-Nezami, H.; Kankaanpaa, P.; Salminen, S.; Ahokas, J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem. Toxicol. 1998, 36, 321–326. [Google Scholar] [CrossRef]
- Gratz, S.; Mykkänen, H.; El-Nezami, H. Aflatoxin B1 binding by a mixture of Lactobacillus and Propionibacterium: In vitro versus ex vivo. J. Food Prot. 2005, 68, 2470–2474. [Google Scholar] [CrossRef] [PubMed]
- Kankaanpää, P.; Tuomola, E.; El-Nezami, H.; Ahokas, J.; Salminen, S.J. Binding of aflatoxin B1 alters the adhesion properties of Lactobacillus rhamnosus strain GG in a Caco-2 model. J. Food Prot. 2000, 63, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Oatley, J.T.; Rarick, M.D.; Ji, G.E.; Linz, J.E. Binding of aflatoxin B1 to Bifidobacteria in vitro. J. Food Prot. 2000, 63, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013, 70, 631–659. [Google Scholar] [CrossRef]
- Tsukita, S.; Furuse, M.; Itoh, M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001, 2, 285–293. [Google Scholar] [CrossRef]
- Tsukita, S.; Furuse, M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann. N. Y. Acad Sci. 2000, 915, 129–135. [Google Scholar] [CrossRef]
- Gao, Y.; Songli, L.; Wang, J.; Luo, C.; Zhao, S.; Zheng, N. Modulation of intestinal epithelial permeability in differentiated Caco-2 cells exposed to aflatoxin M1 and ochratoxin A individually or collectively. Toxins 2018, 10, 13. [Google Scholar] [CrossRef]
- Pandey, I.; Chauhan, S.S. Studies on production performance and toxin residues in tissues and eggs of layer chickens fed on diets with various concentrations of aflatoxin AFB1. Br. Poult. Sci. 2007, 48, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Giambrone, J.J.; Ewert, D.L.; Wyatt, R.D.; Eidson, C.S. Effect of aflatoxin on the humoral and cell-mediated immune systems of the chicken. Am. J. Vet. Res. 1978, 39, 305–308. [Google Scholar] [PubMed]
- Hoerr, F.J. Clinical aspects of immunosuppression in poultry. Avian Dis. 2010, 54, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Yunus, A.W.; Razzazi-Fazeli, E.; Bohm, J. Aflatoxin B1 in affecting broiler’s performance, immunity and gastrointestinal tract: A review of history and contemporary issues. Toxins 2011, 3, 566–590. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.F.; Hamilton, P.B. Impaired phagocytosis by heterophils from chickens during aflatoxicosis. Toxicol. Appl. Pharmacol. 1979, 48, 459–466. [Google Scholar] [CrossRef]
- Chang, C.F.; Hamilton, P.B. Impairment of phagocytosis in chicken monocytes during aflatoxicosis. Poult. Sci. 1979, 58, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.C.; Chauhan, H.V.; Jha, G.J. Suppression of cell-mediated immunity by purified aflatoxin B1 in broiler chicks. Vet. Immunol. Immunopathol. 1991, 28, 165–172. [Google Scholar] [CrossRef]
- Neldon-Ortiz, D.L.; Qureshi, M.A. Effects of AFB1 embryonic exposure on chicken mononuclear phagocytic cell functions. Dev. Comp. Immunol. 1992, 16, 187–196. [Google Scholar] [CrossRef]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.J. Human intestinal barrier function in health and disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef]
- Liu, C.; Zuo, Z.; Zhu, P.; Zheng, Z.; Peng, X.; Cui, H.; Zhou, Y.; Ouyang, P.; Geng, Y.; Deng, J.; et al. Sodium selenite prevents suppression of mucosal humoral response by AFB1 in broiler’s cecal tonsil. Oncotarget 2017, 8, 54215–54226. [Google Scholar]
- Jiang, M.; Fang, J.; Peng, X.; Cui, X.; Yu, Z. Effect of aflatoxin B1 on IgA+ cell number and immunoglobulin mRNA expression in the intestine of broilers. Immunopharmacol. Immunotoxicol. 2015, 37, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yoshinaga, N.; Tanabe, S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J. Biol. Chem. 2011, 286, 31263–31271. [Google Scholar] [CrossRef] [PubMed]
- Kaji, I.; Karaki, S.; Kuwahara, A. Taste sensing in the colon. Curr. Pharm. Des. 2014, 20, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Lee, I.S.; Kim, K.H.; Park, J.; Kim, Y.; Choi, J.H.; Choi, J.S.; Jang, H.J. Activation of intestinal olfactory receptor stimulates glucagon-like peptide-1 secretion in enteroendocrine cells and attenuates hyperglycemia in type 2 diabetic mice. Sci. Rep. 2017, 7, 13978. [Google Scholar] [CrossRef] [PubMed]
- Priori, D.; Colombo, M.; Clavenzani, P.; Jansman, A.J.; Lallès, J.P.; Trevisi, P.; Bosi, P. The olfactory receptor OR51E1 is present along the gastrointestinal tract of pigs, co-localizes with enteroendocrine cells and is modulated by intestinal microbiota. PLoS ONE 2015, 10, e0129501. [Google Scholar] [CrossRef] [PubMed]
- Havenstein, G.B.; Ferket, P.R.; Grimes, J.L.; Qureshi, M.A.; Nestor, K.E. Comparison of the performance of 1966- versus 2003-type turkeys when fed representative 1966 and 2003 turkey diets: Growth rate, livability and feed conversion. Poult. Sci. 2007, 86, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Swalander, M. Aspects of feed efficiency and feeding behaviour in turkeys. Aviagen Turk. Manag. 2015, 4, 9. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]







| Comparison | Groups | Expressed Genes | Shared Genes | Unique Genes (Each Group) | FDR p-Value < 0.05 | |log2FC| > 1.0 | |log2FC| > 2.0 | Up/Down Regulated |
|---|---|---|---|---|---|---|---|---|
| AFB1 | EW (AFB vs. CNTL) | 18744 | 17833 | 402/509 | 703 | 703 | 687 | 674/13 |
| DT (AFB vs. CNTL) | 18654 | 16277 | 304/2073 | 11237 | 7568 | 4515 | 1563/2952 | |
| Line | CNTL (EW vs. DT) | 18736 | 17956 | 386/394 | 679 | 348 | 67 | 37/30 |
| AFB (EW vs. DT) | 18447 | 16369 | 1866/212 | 1666 | 1666 | 1410 | 1308/102 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reed, K.M.; Mendoza, K.M.; Coulombe, R.A., Jr. Differential Transcriptome Responses to Aflatoxin B1 in the Cecal Tonsil of Susceptible and Resistant Turkeys. Toxins 2019, 11, 55. https://doi.org/10.3390/toxins11010055
Reed KM, Mendoza KM, Coulombe RA Jr. Differential Transcriptome Responses to Aflatoxin B1 in the Cecal Tonsil of Susceptible and Resistant Turkeys. Toxins. 2019; 11(1):55. https://doi.org/10.3390/toxins11010055
Chicago/Turabian StyleReed, Kent M., Kristelle M. Mendoza, and Roger A. Coulombe, Jr. 2019. "Differential Transcriptome Responses to Aflatoxin B1 in the Cecal Tonsil of Susceptible and Resistant Turkeys" Toxins 11, no. 1: 55. https://doi.org/10.3390/toxins11010055
APA StyleReed, K. M., Mendoza, K. M., & Coulombe, R. A., Jr. (2019). Differential Transcriptome Responses to Aflatoxin B1 in the Cecal Tonsil of Susceptible and Resistant Turkeys. Toxins, 11(1), 55. https://doi.org/10.3390/toxins11010055