Characterization of Cyanophages in Lake Erie: Interaction Mechanisms and Structural Damage of Toxic Cyanobacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening of Lytic Cyanophages
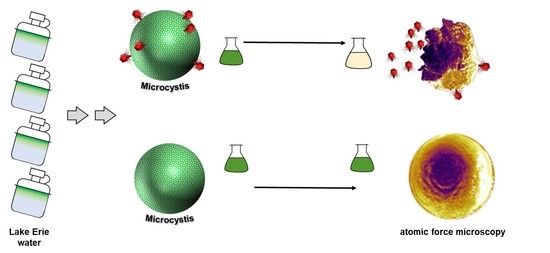
2.2. The Effects of Ma-LEP Infection on Microcystis aeruginosa
2.3. Morphology of Ma-LEP
2.4. Genetic Characterization
3. Materials and Methods
3.1. Water Sample Collection, Concentration, and Screening of Lytic Cyanophages
3.2. Host Bacteria: Microcystis aeruginosa
3.3. The Effects of Cyanophage Infections on Microcystis aeruginosa
3.4. Atomic Force Microscopy
3.5. TEM
3.6. Targeting Viral Genes Using PCR
3.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davis, T.W.; Gobler, C.J. Preface for Special Issue on “Global expansion of harmful cyanobacterial blooms: Diversity, ecology, causes, and controls”. Harmful Algae 2016, 54, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, P.; Anderson, D.M.; Kaoru, Y.; White, A.W. The economic effects of harmful algal blooms in the United States: Estimates, assessment issues, and information needs. Estuaries 2002, 25, 819–837. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Jiang, X. Cyanobacterial Toxins in Freshwater and Food: Important Sources of Exposure to Humans. Annu. Rev. Food Sci. 2017, 8, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.; Campos, A.; Vasconcelos, V.; Freitas, M. Effects of microcystin-LR and cylindrospermopsin on plant-soil systems: A review of their relevance for agricultural plant quality and public health. Environ. Res. 2016, 153, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Bin Alam, M.Z.; Otaki, M.; Furumai, H.; Ohgaki, S. Direct and indirect inactivation of Microcystis aeruginosa by UV-radiation. Water Res. 2001, 35, 1008–1014. [Google Scholar] [CrossRef]
- Yu, Z.; Song, X.; Cao, X.; Liu, Y. Mitigation of harmful algal blooms using modified clays: Theory, mechanisms, and applications. Harmful Algae 2017, 69, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Q.; Wei, Z.; Liu, N.; Li, Y.; Li, D.; Jin, Z.; Xu, X. Thiazole Amides, A Novel Class of Algaecides against Freshwater Harmful Algae. Sci. Rep. 2018, 8, 8555. [Google Scholar] [CrossRef] [PubMed]
- Mitsch, W.J. Solving Lake Erie’s harmful algal blooms by restoring the Great Black Swamp in Ohio. Ecol. Eng. 2017, 108, 406–413. [Google Scholar] [CrossRef]
- Paerl, H.W.; Otten, T.G.; Kudela, R. Mitigating the Expansion of Harmful Algal Blooms Across the Freshwater-to-Marine Continuum. Environ. Sci. Technol. 2018, 52, 5519–5529. [Google Scholar] [CrossRef] [PubMed]
- Mann, N.H. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 2003, 27, 17–34. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Sullivan, M.B.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses Ocean microbes drive biogeochemical cycling on a global scale. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, T.; Deng, F.; Hu, Z. Freshwater cyanophages. Virol. Sin. 2013, 28, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Yoshida, T.; Kashima, A.; Takashima, Y.; Hosoda, N.; Nagasaki, K.; Hiroishi, S. Ecological dynamics of the toxic bloom-forming cyanobacterium Microcystis aeruginosa and its cyanophages in freshwater. Appl. Environ. Microbiol. 2008, 74, 3269–3273. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Yoshida, T.; Yoshida-Takashima, Y.; Kashima, A.; Hiroishi, S. Real-time PCR detection of host-mediated cyanophage gene transcripts during infection of a natural Microcystis aeruginosa population. Microbes Environ. 2010, 25, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Wang, M.; Ge, X.; Wu, Y.; Yang, X.; Zhang, Y.; Li, T.; Shi, Z. Study of the dynamics of Microcystis aeruginosa and its cyanophage in East Lake using quantitative PCR. Virol. Sin. 2013, 28, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.Y.; Liang, S.; Lee, J. Toxin-producing cyanobacteria in freshwater: A review of the problems, impact on drinking water safety, and efforts for protecting public health. J. Microbiol. 2013, 51, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Van Wichelen, J.; Vanormelingen, P.; Codd, G.A.; Vyverman, W. The common bloom-forming cyanobacterium Microcystis is prone to a wide array of microbial antagonists. Harmful Algae 2016, 55, 97–111. [Google Scholar] [CrossRef]
- Meaden, S.; Koskella, B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013, 4, 358. [Google Scholar] [CrossRef] [Green Version]
- Doss, J.; Culbertson, K.; Hahn, D.; Camacho, J.; Barekzi, N. A Review of Phage Therapy against Bacterial Pathogens of Aquatic and Terrestrial Organisms. Viruses 2017, 9, 50. [Google Scholar] [CrossRef]
- Ou, T.; Gao, X.C.; Li, S.H.; Zhang, Q.Y. Genome analysis and gene nblA identification of microcystis aeruginosa myovirus (MaMV-DC) reveal the evidence for horizontal gene transfer events between cyanomyovirus and host. J. Gen. Virol. 2015, 96, 3681–3697. [Google Scholar] [CrossRef]
- Gao, E.-B.; Gui, J.-F.; Zhang, Q.-Y. A novel cyanophage with a cyanobacterial nonbleaching protein A gene in the genome. J. Virol. 2012, 86, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Safferman, R.; Morris, M. Growth characteristics of the blue-green algal virus LPP-1. J. Bacteriol. 1964, 88, 771–775. [Google Scholar] [PubMed]
- Tucker, S.; Pollard, P. Identification of cyanophage Ma-LBP and infection of the cyanobacterium Microcystis aeruginosa from an Australian subtropical lake by the virus. Appl. Environ. Microbiol. 2005, 71, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Takashima, Y.; Tomaru, Y.; Takao, Y.; Hiroishi, S.; Shirai, Y. Isolation and Characterization of a Cyanophage Infecting the Toxic Cyanobacterium Microcystis aeruginosa Isolation and Characterization of a Cyanophage Infecting the Toxic Cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 2006, 72, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Liao, X.-Y.; Gao, X.-C.; Xu, X.-D.; Zhang, Q.-Y. Unraveling the genome structure of cyanobacterial podovirus A-4L with long direct terminal repeats. Virus Res. 2015, 203, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Short, C.M.; Suttle, C.A. Nearly Identical Bacteriophage Structural Gene Sequences Are Widely Distributed in both Marine and Freshwater Environments Nearly Identical Bacteriophage Structural Gene Sequences Are Widely Distributed in both Marine and Freshwater Environments. Appl. Environ. Microbiol. 2005, 71, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Mohiuddin, M.; Schellhorn, H.E. Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis. Front. Microbiol. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kimura-Sakai, S.; Sako, Y.; Yoshida, T. Development of a real-time PCR assay for the quantification of Ma-LMM01-type Microcystis cyanophages in a natural pond. Lett. Appl. Microbiol. 2015, 60, 400–408. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Cowan, D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl. Environ. Microbiol. 2014, 80, 4470–4480. [Google Scholar] [CrossRef]
- Baclayon, M.; Wuite, G.J.L.; Roos, W.H. Imaging and manipulation of single viruses by atomic force microscopy. Soft Matter 2010, 6, 5273. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; McPherson, A. Atomic Force Microscopy in Imaging of Viruses and Virus-Infected Cells. Microbiol. Mol. Biol. Rev. 2011, 75, 268–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, S.C.; Smith, J.R.; Hayes, P.K.; Watts, J.E.M. Characterisation of host growth after infection with a broad-range freshwater cyanopodophage. PLoS ONE 2014, 9, e87339. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.; Clokie, M.R.J.; Millard, A.; Mann, N.H. Cyanophage infection and photoinhibition in marine cyanobacteria. Res. Microbiol. 2004, 155, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.J.; Mann, N.H. Marine cyanophages and light. Environ. Microbiol. 2006, 8, 2074–2082. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Prangishvili, D.; Lindell, D. Roles of viruses in the environment. Environ. Microbiol. 2009, 11, 2771–2774. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.H. The Origin of Oxygenic Photosynthesis and Its Impact on the Atmosphere. In Earth’s Early Atmosphere and Oceans, and The Origin of Life; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 75–93. [Google Scholar]
- Honda, T.; Takahashi, H.; Sako, Y.; Yoshida, T. Gene expression of Microcystis aeruginosa during infection of cyanomyovirus Ma-LMM01. Fish. Sci. 2014, 80, 83–91. [Google Scholar] [CrossRef]
- Haramoto, E.; Katayama, H.; Oguma, K.; Ohgaki, S. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 2005, 71, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Shirai, M.; Matumaru, K.; Ohotake, A.; Takamura, Y.; Aida, T.; Nakano, M. Development of a Solid Medium for Growth and Isolation of Axenic Microcystis Strains (Cyanobacteria). Appl. Environ. Microbiol. 1989, 55, 2569. [Google Scholar]
- Jiang, X.; Lee, S.; Mok, C.; Lee, J. Sustainable Methods for Decontamination of Microcystin in Water Using Cold Plasma and UV with Reusable TiO2 Nanoparticle Coating. Int. J. Environ. Res. Public Health 2017, 14, 480. [Google Scholar] [CrossRef]
- Schmidt, K.; Mwaigwisya, S.; Crossman, L.C.; Doumith, M.; Munroe, D.; Pires, C.; Khan, A.M.; Woodford, N.; Saunders, N.J.; Wain, J.; et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 2017, 72, 104–114. [Google Scholar] [CrossRef]
- Quick, J.; Ashton, P.; Calus, S.; Chatt, C.; Gossain, S.; Hawker, J.; Nair, S.; Neal, K.; Nye, K.; Peters, T.; et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 2015, 16, 114. [Google Scholar] [CrossRef]
- Marion, J.W.; Lee, J.; Wilkins, J.R.; Lemeshow, S.; Lee, C.; Waletzko, E.J.; Buckley, T.J. In vivo phycocyanin flourometry as a potential rapid screening tool for predicting elevated microcystin concentrations at eutrophic lakes. Environ. Sci. Technol. 2012, 46, 4523–4531. [Google Scholar] [CrossRef] [PubMed]
- US EPA. Method 546: Determination of Total Microcystins and Nodularins in Drinking Water and Ambient Water by Adda Enzyme-Linked Immunosorbent Assay; EPA: Washington, DC, USA, 2016.
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Dekel-Bird, N.P.; Avrani, S.; Sabehi, G.; Pekarsky, I.; Marston, M.F.; Kirzner, S.; Lindell, D. Diversity and evolutionary relationships of T 7-like podoviruses infecting marine cyanobacteria. Environ. Microbiol. 2013, 15, 1476–1491. [Google Scholar]





| Treatment | Phycocyanin | Chlorophyll-a | OD at 680 nm | Log (Gene Copy Number) 1 | ||||
|---|---|---|---|---|---|---|---|---|
| Slope | R2 | Slope | R2 | Slope | R2 | Slope | R2 | |
| Control | 13.10 | 0.93 | 3.45 | 0.84 | 0.04 | >0.99 | 0.28 | 0.82 |
| Cyanophage Ma-LEP | 5.75 ** | 0.68 | 2.18 ** | 0.90 | 0.03 ** | 0.99 | 0.13 ** | 0.74 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Ha, C.; Lee, S.; Kwon, J.; Cho, H.; Gorham, T.; Lee, J. Characterization of Cyanophages in Lake Erie: Interaction Mechanisms and Structural Damage of Toxic Cyanobacteria. Toxins 2019, 11, 444. https://doi.org/10.3390/toxins11080444
Jiang X, Ha C, Lee S, Kwon J, Cho H, Gorham T, Lee J. Characterization of Cyanophages in Lake Erie: Interaction Mechanisms and Structural Damage of Toxic Cyanobacteria. Toxins. 2019; 11(8):444. https://doi.org/10.3390/toxins11080444
Chicago/Turabian StyleJiang, Xuewen, Chanhee Ha, Seungjun Lee, Jinha Kwon, Hanna Cho, Tyler Gorham, and Jiyoung Lee. 2019. "Characterization of Cyanophages in Lake Erie: Interaction Mechanisms and Structural Damage of Toxic Cyanobacteria" Toxins 11, no. 8: 444. https://doi.org/10.3390/toxins11080444
APA StyleJiang, X., Ha, C., Lee, S., Kwon, J., Cho, H., Gorham, T., & Lee, J. (2019). Characterization of Cyanophages in Lake Erie: Interaction Mechanisms and Structural Damage of Toxic Cyanobacteria. Toxins, 11(8), 444. https://doi.org/10.3390/toxins11080444