Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins
Abstract
:1. Introduction
2. Results
2.1. Bacterial Growth in the Presence of Mycotoxins
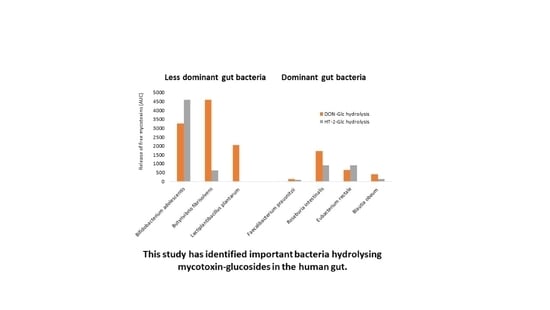
2.2. Hydrolysis of Masked Trichothecenes by Bacterial Strains
2.3. Time-Course of Hydrolysis of Masked Trichothecenes by Selected Bacterial Strains
2.4. Degradation of Unconjugated Mycotoxins by Bacterial Strains
2.4.1. De-Epoxydation of Trichothecenes
2.4.2. De-Acetylation of Type A Trichothecenes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Medium Selection
4.3. Mycotoxin Standards
4.4. Bacterial Growth and Mycotoxin Metabolism in Anaerobic 96-Well Plates over 48 h
4.5. Time Course Experiments of Mycotoxin Metabolism of Selected Bacterial Strains
4.6. LC-MS/MS Analysis
4.7. Statistical Analysis
- Bacterial control vs. solvent control a
- Bacterial control vs. solvent control b
- Solvent control a vs. mycotoxin a
- Solvent control b vs. mycotoxin b
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, H.J.; Ryu, D. Worldwide Occurrence of Mycotoxins in Cereals and Cereal-Derived Food Products: Public Health Perspectives of Their Co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef] [PubMed]
- Bryla, M.; Waskiewicz, A.; Ksieniewicz-Wozniak, E.; Szymczyk, K.; Jedrzejczak, R. Modified Fusarium Mycotoxins in Cereals and their Products-Metabolism, Occurrence, and Toxicity: An Updated Review. Molecules 2018, 23, 963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broekaert, N.; Devreese, M.; De Baere, S.; De Backer, P.; Croubels, S. Modified Fusarium mycotoxins unmasked: From occurrence in cereals to animal and human excretion. Food Chem. Toxicol. 2015, 80, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; De Saeger, S.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked Mycotoxins: A Review. Mol. Nutr. Food Res. 2013, 57, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.W. Do Plant-Bound Masked Mycotoxins Contribute to Toxicity? Toxins 2017, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Dall’Erta, A.; Cirlini, M.; Dall’Asta, M.; Del Rio, D.; Galaverna, G.; Dall’Asta, C. Masked Mycotoxins Are Efficiently Hydrolyzed by Human Colonic Microbiota Releasing Their Aglycones. Chem. Res. Toxicol. 2013, 26, 305–312. [Google Scholar] [CrossRef]
- Daud, N.; Currie, V.; Duncan, G.; Busman, M.; Gratz, S.W. Intestinal Hydrolysis and Microbial Biotransformation of Diacetoxyscirpenol-Alpha-Glucoside, HT-2-Beta-Glucoside and N-(1-Deoxy-D-Fructos-1-Yl) Fumonisin B-1 by Human Gut Microbiota in Vitro. Int. J. Food Sci. Nutr. 2020, 71, 540–548. [Google Scholar] [CrossRef]
- Gratz, S.W.; Duncan, G.; Richardson, A.J. The Human Fecal Microbiota Metabolizes Deoxynivalenol and Deoxynivalenol-3-Glucoside and May Be Responsible for Urinary Deepoxy-Deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1821–1825. [Google Scholar] [CrossRef] [Green Version]
- Gratz, S.W.; Dinesh, R.; Yoshinari, T.; Holtrop, G.; Richardson, A.J.; Duncan, G.; Macdonald, S.; Lloyd, A.; Tarbin, J. Masked trichothecene and zearalenone mycotoxins withstand digestion and absorption in the upper GI tract but are efficiently hydrolyzed by human gut microbiota in vitro. Mol. Nutr. Food Res. 2017, 61, 1600680. [Google Scholar] [CrossRef]
- McCormick, S.P.; Kato, T.; Maragos, C.; Busman, M.; Lattanzio, V.M.T.; Galaverna, G.; Dall’Asta, C.; Crich, D.; Price, N.P.J.; Kurtzman, C.P. Anomericity of T-2 Toxin-glucoside: Masked Mycotoxin in Cereal Crops. J. Agric. Food Chem. 2015, 63, 731–738. [Google Scholar] [CrossRef]
- Eriksen, G.S.; Pettersson, H.; Johnsen, K.; Lindberg, J.E. Transformation of Trichothecenes in Ileal Digesta and Faeces from Pigs. Arch. Anim. Nutr. 2002, 56, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.W.; Currie, V.; Richardson, A.J.; Duncan, G.; Holtrop, G.; Farquharson, F.; Louis, P.; Pinton, P.; Oswald, I.P. Porcine Small and Large Intestinal Microbiota Rapidly Hydrolyze the Masked Mycotoxin Deoxynivalenol-3-Glucoside and Release Deoxynivalenol in Spiked Batch Cultures In Vitro. Appl. Environ. Microbiol. 2018, 84, e02106-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, A.; Claeys, L.; Mengelers, M.; Vanhoorne, V.; Vervaet, C.; Huybrechts, B.; De Saeger, S.; De Boevre, M. Humans significantly metabolize and excrete the mycotoxin deoxynivalenol and its modified form deoxynivalenol-3-glucoside within 24 hours. Sci. Rep. 2018, 8, 5255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M. Enterotypes of the human gut microbiome. Nat. Cell Biol. 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Ramakrishna, B. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef]
- Jandhyala, S.M. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Michlmayr, H.; Varga, E.; Malachova, A.; Nhung, T.N.; Lorenz, C.; Haltrich, D.; Berthiller, F.; Adam, G. A Versatile Family 3 Glycoside Hydrolase from Bifidobacterium Adolescentis Hydrolyzes Beta-Glucosides of the Fusarium Mycotoxins Deoxynivalenol, Nivalenol, and HT-2 Toxin in Cereal Matrices. Appl. Environ. Microbiol. 2015, 81, 4885–4893. [Google Scholar] [CrossRef] [Green Version]
- Le Sciellour, M.; Zemb, O.; Serviento, A.-M.; Renaudeau, D. Transient effect of single or repeated acute deoxynivalenol and zearalenone dietary challenge on fecal microbiota composition in female finishing pigs. Animal 2020, 1–11. [Google Scholar] [CrossRef]
- Miró-Abella, E.; Torrell, H.; Herrero, P.; Canela, N.; Arola, L.; Borrull, F.; Ras, R.; Fontanals, N. Monitoring and evaluation of the interaction between deoxynivalenol and gut microbiota in Wistar rats by mass spectrometry-based metabolomics and next-generation sequencing. Food Chem. Toxicol. 2018, 121, 124–130. [Google Scholar] [CrossRef]
- Reddy, K.E.; Jeong, J.; Song, J.; Lee, Y.K.; Lee, H.-J.; Kim, D.-W.; Jung, H.J.; Kim, K.H.; Kim, M.; Oh, Y.K.; et al. Colon Microbiome of Pigs Fed Diet Contaminated with Commercial Purified Deoxynivalenol and Zearalenone. Toxins 2018, 10, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; De Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcenilla, A.; Pryde, S.E.; Martin, J.C.; Duncan, S.H.; Stewart, C.S.; Henderson, C.; Flint, H.J. Phylogenetic Relationships of Butyrate-Producing Bacteria from the Human Gut. Appl. Environ. Microbiol. 2000, 66, 1654–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellani, A.; Chalmers, A. Manual of Tropical Medicine, 3rd ed.; Williams Wood and Co.: New York, NY, USA, 1919. [Google Scholar] [CrossRef] [Green Version]
- Hahnke, R.L.; Meier-Kolthoff, J.P.; García-López, M.; Mukherjee, S.; Huntemann, M.; Ivanova, N.N.; Woyke, T.; Kyrpides, N.C.; Klenk, H.-P.; Göker, M. Genome-Based Taxonomic Classification of Bacteroidetes. Front. Microbiol. 2016, 7, 2003. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, H.; Shibata, K.; Sakamoto, M.; Tomita, S.; Benno, Y. Prevotella Copri Sp Nov and Prevotella Stercorea Sp Nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2007, 57, 941–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcenilla, A. Diversity of the Butyrate-Producing Microflora of the Human Gut. Ph.D. Thesis, Robert Gordon University, Aberdeen, UK, 1999. [Google Scholar]
- Holdeman, L.V.; Moore, W.E.C. New Genus, Coprococcus, Twelve New Species, and Emended Descriptions of Four Previously Described Species of Bacteria from Human Feces. Int. J. Syst. Bacteriol. 1974, 24, 260–277. [Google Scholar] [CrossRef] [Green Version]
- Shetty, S.A.; Zuffa, S.; Bui, T.P.N.; Aalvink, S.; Smidt, H.; De Vos, W.M. Reclassification of Eubacterium hallii as Anaerobutyricum hallii gen. nov., comb. nov., and description of Anaerobutyricum soehngenii sp. nov., a butyrate and propionate-producing bacterium from infant faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 3741–3746. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted Distribution of the Butyrate Kinase Pathway among Butyrate-Producing Bacteria from the Human Colon. J. Bacteriol. 2004, 186, 2099–2106. [Google Scholar] [CrossRef] [Green Version]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef]
- Rumney, C.; Duncan, S.; Henderson, C.; Stewart, C. Isolation and Characteristics of a Wheatbran-Degrading Butyrivibrio from Human Feces. Lett. Appl. Microbiol. 1995, 20, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Alessi, A.M.; Gray, V.; Farquharson, F.M.; Flores-Lopez, A.; Shaw, S.; Stead, D.; Wegmann, U.; Shearman, C.; Gasson, M.; Collie-Duguid, E.S.R.; et al. Beta-Glucan is a Major Growth Substrate for Human Gut Bacteria Related to Coprococcus Eutactus. Environ. Microbiol. 2020, 22, 2150–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.; Hold, G.; Barcenilla, A.; Stewart, C.; Flint, H. Roseburia Intestinalis Sp Nov., a Novel Saccharolytic, Butyrate-Producing Bacterium from Human Faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 1615–1620. [Google Scholar] [PubMed] [Green Version]
- Sharpe, M.E. A Serological Classification of Lactobacilli. J. Gen. Microbiol. 1955, 12, 107–122. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.; Farrow, J.; Jones, D. Enterococcus-Mundtii Sp-Nov. Int. J. Syst. Bacteriol. 1986, 36, 8–12. [Google Scholar] [CrossRef] [Green Version]
- El-Nezami, H.; Polychronaki, N.; Salminen, S.; Mykkanen, H. Binding rather than Metabolism may Explain the Interaction of Two Food-Grade Lactobacillus Strains with Zearalenone and its Derivative Alpha-Zearalenol. Appl. Environ. Microbiol. 2002, 68, 3545–3549. [Google Scholar] [CrossRef] [Green Version]
- El-Nezami, H.; Chrevatidis, A.; Auriola, S.; Salminen, S.; Mykkänen, H. Removal of common Fusarium toxins in vitro by strains of Lactobacillus and Propionibacterium. Food Addit. Contam. 2002, 19, 680–686. [Google Scholar] [CrossRef]
- Fuchs, S.; Sontag, G.; Stidl, R.; Ehrlich, V.; Kundi, M.; Knasmüller, S. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008, 46, 1398–1407. [Google Scholar] [CrossRef]
- Berthiller, F.; Krska, R.; Domig, K.J.; Kneifel, W.; Juge, N.; Schuhmacher, R.; Adam, G. Hydrolytic fate of deoxynivalenol-3-glucoside during digestion. Toxicol. Lett. 2011, 206, 264–267. [Google Scholar] [CrossRef] [Green Version]
- McCormick, S.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From Simple to Complex Mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef]
- Aminov, R.I.; Walker, A.W.; Duncan, S.H.; Harmsen, H.J.M.; Welling, G.W.; Flint, H.J. Molecular Diversity, Cultivation, and Improved Detection by Fluorescent In Situ Hybridization of a Dominant Group of Human Gut Bacteria Related to Roseburia spp. or Eubacterium rectale. Appl. Environ. Microbiol. 2006, 72, 6371–6376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, O.; Knights, D.; Gonzalez, A.; Waldron, L.; Segata, N.; Knight, R.; Huttenhower, C.; Ley, R.E. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS Comput. Biol. 2013, 9, e1002863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2008, 101, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeney, D.D.; Gareau, M.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gratz, S.W.; Currie, V.; Duncan, G.; Jackson, D. Multimycotoxin Exposure Assessment in UK Children Using Urinary Biomarkers—A Pilot Survey. J. Agric. Food Chem. 2019, 68, 351–357. [Google Scholar] [CrossRef]
- Schothorst, R.C.; Van Egmond, H.P. Report from SCOOP task 3.2.10 “collection of occurrence data of Fusarium toxins in food and assessment of dietary intake by the population of EU member states”. Toxicol. Lett. 2004, 153, 133–143. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Cotta, M.A.; Hespell, R.B. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 1986, 52, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.-P.F.L.; Schatzmayr, G.; He, J.W.; Zhou, T.; Moll, W.-D.; et al. Microbial biotransformation of DON: Molecular basis for reduced toxicity. Sci. Rep. 2016, 6, 29105. [Google Scholar] [CrossRef] [Green Version]
- Madhyastha, M.; Marquardt, R.; Abramson, D. Structure-activity relationships and interactions among trichothecene mycotoxins as assessed by yeast bioassay. Toxicon 1994, 32, 1147–1152. [Google Scholar] [CrossRef]
- Wu, Q.; Dohnal, V.; Kuca, K.; Yuan, Z. Trichothecenes: Structure-Toxic Activity Relationships. Curr. Drug Metab. 2013, 14, 641–660. [Google Scholar] [CrossRef] [PubMed]
- Behm, C.; Föllmann, W.; Degen, G.H. Cytotoxic Potency of Mycotoxins in Cultures of V79 Lung Fibroblast Cells. J. Toxicol. Environ. Health Part A 2012, 75, 1226–1231. [Google Scholar] [CrossRef]
- Tran, V.; Viktorová, J.; Augustynkova, K.; Jelenova, N.; Dobiasová, S.; Řehořová, K.; Fenclová, M.; Stranska-Zachariasova, M.; Vítek, L.; HajšLová, J.; et al. In Silico and In Vitro Studies of Mycotoxins and Their Cocktails; Their Toxicity and Its Mitigation by Silibinin Pre-Treatment. Toxins 2020, 12, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella copri Isolates. Cell Host Microbe 2019, 26, 680–690.e5. [Google Scholar] [CrossRef]
- Fragiadakis, G.K.; Smits, S.A.; Sonnenburg, E.D.; Van Treuren, W.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Dominguez-Bello, M.G.; Leach, J.; et al. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 2018, 10, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E.; et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef] [Green Version]
- Ruengsomwong, S.; La-Ongkham, O.; Jiang, J.; Wannissorn, B.; Nakayama, J.; Nitisinprasert, S. Microbial Community of Healthy Thai Vegetarians and Non-Vegetarians, Their Core Gut Microbiota, and Pathogen Risk. J. Microbiol. Biotechnol. 2016, 26, 1723–1735. [Google Scholar] [CrossRef] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Matsushima, T.; Okamoto, E.; Miyagawa, E.; Matsui, Y.; Shimizu, H.; Asano, K. Deacetylation of diacetoxyscirpenol to 15-acetoxyscirpenol by rumen bacteria. J. Gen. Appl. Microbiol. 1996, 42, 225–234. [Google Scholar] [CrossRef] [Green Version]
- Robert, H.; Payros, D.; Pinton, P.; Théodorou, V.; Mercier-Bonin, M.; Oswald, I.P. Impact of mycotoxins on the intestine: Are mucus and microbiota new targets? J. Toxicol. Environ. Health Part B 2017, 20, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.; Hold, G.; Harmsen, H.; Stewart, C.; Flint, H. Growth Requirements and Fermentation Products of Fusobacterium Prausnitzii, and a Proposal to Reclassify it as Faecalibacterium Prausnitzii Gen. Nov., Comb. Nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [PubMed] [Green Version]
- Hassan, F.-U.; Ebeid, H.M.; Tang, Z.; Li, M.; Peng, L.; Peng, K.; Liang, X.; Yang, C. A Mixed Phytogenic Modulates the Rumen Bacteria Composition and Milk Fatty Acid Profile of Water Buffaloes. Front. Veter. Sci. 2020, 7, 569. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Price, N.P.J.; Kurtzman, C.P. Glucosylation and Other Biotransformations of T-2 Toxin by Yeasts of the Trichomonascus Clade. Appl. Environ. Microbiol. 2012, 78, 8694–8702. [Google Scholar] [CrossRef] [Green Version]
- Wetterhorn, K.M.; Newmister, S.A.; Caniza, R.K.; Busman, M.; McCormick, S.P.; Berthiller, F.; Adam, G.; Rayment, I. Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol. Biochemestry 2016, 55, 6175–6186. [Google Scholar] [CrossRef]
- Yoshinari, T.; Sakuda, S.; Furihata, K.; Furusawa, H.; Ohnishi, T.; Sugita-Konishi, Y.; Ishizaki, N.; Terajima, J. Structural Determination of a Nivalenol Glucoside and Development of an Analytical Method for the Simultaneous Determination of Nivalenol and Deoxynivalenol, and Their Glucosides, in Wheat. J. Agric. Food Chem. 2014, 62, 1174–1180. [Google Scholar] [CrossRef]
- Soto-Martin, E.C.; Warnke, I.; Farquharson, F.M.; Christodoulou, M.; Horgan, G.; Derrien, M.; Faurie, J.-M.; Flint, H.J.; Duncan, S.H.; Louis, P. Vitamin Biosynthesis by Human Gut Butyrate-Producing Bacteria and Cross-Feeding in Synthetic Microbial Communities. mBio 2020, 11. [Google Scholar] [CrossRef]
- Bretz, F.; Hothorn, T.; Westfall, P. Multiple Comparisons Using R; Informa UK Limited: London, UK, 2016; p. 205. [Google Scholar]

| Compound Name | Abbreviation | R1 | R2 | R3 | R4 | R5 |
|---|---|---|---|---|---|---|
| T-2 toxin | T-2 | OH | OAc | OAc | H | C5H9O2 |
| HT-2 toxin | HT-2 | OH | OH | OAc | H | C5H9O2 |
| T-2-3-α,D-glucoside | T-2-Glc | C6H11O6 | OAc | OAc | H | C5H9O2 |
| HT-2-3-β,D-glucoside | HT-2-Glc | C6H11O6 | OH | OAc | H | C5H9O2 |
| 4,15-Diacetoxyscirpenol | DAS | OH | OAc | OAc | H | H |
| 15-Monoacetoxyscirpenol | 15-MAS | OH | OH | OAc | H | H |
| 4-Monoacetoxyscirpenol | 4-MAS | OH | OAc | OH | H | H |
| Scirpenol | SCP | OH | OH | OH | H | H |
| DAS-3-α,D-glucoside | DAS-Glc | C6H11O6 | OAc | OAc | H | H |
| Deoxynivalenol | DON | OH | H | OH | OH | =O |
| DON-3-β,D-glucoside | DON-Glc | C6H11O6 | H | OH | OH | =O |
| Nivalenol | NIV | OH | OH | OH | OH | =O |
| NIV-3- β,D-glucoside | NIV-Glc | C6H11O6 | OH | OH | OH | =O |







| Phylum | Family | Bacterial Species | Reference(s) |
|---|---|---|---|
| Verrucomicrobia | Akkermansiaceae | Akkermansia muciniphila DSM 22959 | [22] |
| Actinobacteria | Bifidobacteriaceae | Bifidobacterium adolescentis DSM 20083 | [23] |
| Bacteroidetes | Bacteroidaceae | Bacteroides thetaiotaomicron DSM 2079 | [24,25] |
| Bacteroidetes | Bacteroidaceae | Prevotella copri DSM 18205 | [26] |
| Firmicutes | Erysipelotrichaceae | Faecalitalea cylindroides T2-87 | [27] |
| Firmicutes | Lachnospiraceae | Anaerobutyricum hallii DSM 3353 | [28,29] |
| Firmicutes | Lachnospiraceae | Anaerostipes hadrus SSC/2 | [30] |
| Firmicutes | Lachnospiraceae | Blautia obeum A2-162 | [27,31,32] |
| Firmicutes | Lachnospiraceae | Butyrivibrio fibrisolvens 16/4 | [33] |
| Firmicutes | Lachnospiraceae | Coprococcus sp. ART55/1 | [30,34] |
| Firmicutes | Lachnospiraceae | Eubacterium rectale DSM 17629 (A1-86) | [23] |
| Firmicutes | Lachnospiraceae | Roseburia intestinalis L1-82 | [23,35] |
| Firmicutes | Lactobacillaceae | Lactiplantibacillus plantarum NCIMB 7220 | [36] |
| Firmicutes | Ruminococcaceae | Faecalibacterium prausnitzii A2-165 | [23] |
| Firmicutes | Enterococcaceae | Enterococcus mundtii DSM 4838 * | [37] |
| Bacterial Strain | Bacterial | Solvent Control (nmol/mL) | DON (nmol/mL) | DON-Glc (nmol/mL) | HT-2 (nmol/mL) | HT-2-Glc (nmol/mL) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 2 | 10 | 2 | 10 | 2 | 10 | 2 | 10 | 2 | 10 | |
| A. muciniphila DSM 22959 | 0.148 | 0.197 | 0.191 | 0.210 | 0.202 | 0.131 | 0.141 | 0.128 | 0.127 | 0.122 | 0.150 |
| B. adolescentis DSM 20083 | 0.747 | 0.708 | 0.732 | 0.728 | 0.715 | 0.619 | 0.622 | 0.764 | 0.656 | 0.642 | 0.647 |
| B. thetaiotaomicron DSM 2079 | 0.099 | 0.157 | 0.107 | 0.085 | 0.108 | 0.110 | 0.100 | 0.097 | 0.089 | 0.121 | 0.096 |
| P. copri DSM 18205 | 0.367 | 0.355 | 0.343 | 0.366 | 0.384 | 0.364 | 0.375 | 0.368 | 0.363 | 0.377 | 0.376 |
| F. cylindroides T2-87 | 0.570 | 0.536 | 0.548 | 0.629 | 0.628 | 0.552 | 0.665 | 0.574 | 0.582 | 0.501 | 0.576 |
| A. hallii DSM 3353 | 1.072 | 1.197 | 1.328 | 1.017 | 1.123 | 1.511 | 0.964 | 1.281 | 1.213 | 1.285 | 1.295 |
| A. hadrus SSC/2 | 1.111 | 1.170 | 1.074 | 1.055 | 1.113 | 1.009 | 1.180 | 1.141 | 1.061 | 1.147 | 1.164 |
| B. obeum A2-162 | 0.240 | 0.260 | 0.243 | 0.240 | 0.243 | 0.235 | 0.261 | 0.251 | 0.264 | 0.255 | 0.268 |
| B. fibrisolvens 16/4 | 0.254 | 0.237 | 0.317 | 0.220 | 0.290 | 0.265 | 0.292 | 0.271 | 0.289 | 0.298 | 0.367 |
| Coprococcus sp. ART55/1 | 0.775 | 0.834 | 0.852 | 0.782 | 0.803 | 0.696 | 0.941 | 0.781 | 0.678 | 0.626 | 0.551 |
| E. rectale DSM 17629 | 0.301 | 0.308 | 0.328 | 0.290 | 0.333 | 0.317 | 0.340 | 0.337 | 0.357 | 0.345 | 0.358 |
| R. intestinalis L1-82 | 1.229 | 1.178 | 1.141 | 1.208 | 1.188 | 1.154 | 1.153 | 1.156 | 1.172 | 1.222 | 1.120 |
| L. plantarum NCIMB 7220 | 0.731 | 0.733 | 0.747 | 0.768 | 0.800 | 0.784 | 0.769 | 0.772 | 0.765 | 0.773 | 0.781 |
| F. prausnitzii A2-165 | 0.523 | 0.552 | 0.531 | 0.559 | 0.526 | 0.539 | 0.546 | 0.523 | 0.503 | 0.500 | 0.453 |
| E. mundtii DSM 4838 | 1.325 | 1.286 | 1.379 | 1.273 | 1.381 | 1.560 | 1.361 | 1.371 | 1.347 | 1.281 | 1.286 |
| Compound | Precursor Ion (m/z) | Product Ion (m/z) | Retention Time (RT) (min) | Dwell Time (msec) | Collision Energy | Polarity |
|---|---|---|---|---|---|---|
| DOM-1 | 339.1 | 249.10 | 11.64 | 75 | −16.0 | Negative |
| DON | 355.1 | 265.10 | 9.47 | 75 | −21.0 | Negative |
| DON-Glc | 517.3 | 427.30 | 8.65 | 75 | −29.0 | Negative |
| NIV | 371.1 | 281.1 | 6.76 | 75 | −21.5 | Negative |
| NIV-Glc | 533.3 | 473.4 | 6.16 | 75 | −19.5 | Negative |
| deNIV | 355.1 | 265.1 | 6.96 | 75 | −21.5 | Negative |
| T-2 | 484.4 | 185.3 | 12.87 | 50 | 30.5 | Positive |
| T-2-Glc | 646.4 | 305.1 | 11.76 | 50 | 26.5 | Positive |
| HT-2 | 442.3 | 215.3 | 11.60 | 50 | 17.0 | Positive |
| HT-2-Glc | 604.4 | 323.1 | 10.53 | 50 | 17.0 | Positive |
| DAS | 384.4 | 307.3 | 9.31 | 50 | 17.0 | Positive |
| DAS-Glc | 546.3 | 349.3 | 7.99 | 50 | 20.5 | Positive |
| 15-MAS | 342.2 | 265.2 | 7.80 | 50 | 12.0 | Positive |
| 4-MAS | 342.2 | 217.1 | 5.90 | 50 | 17.0 | Positive |
| SCP | 300.2 | 247.1 | 5.3 | 50 | 17.0 | Positive |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daud, N.; Currie, V.; Duncan, G.; Farquharson, F.; Yoshinari, T.; Louis, P.; Gratz, S.W. Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins. Toxins 2020, 12, 654. https://doi.org/10.3390/toxins12100654
Daud N, Currie V, Duncan G, Farquharson F, Yoshinari T, Louis P, Gratz SW. Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins. Toxins. 2020; 12(10):654. https://doi.org/10.3390/toxins12100654
Chicago/Turabian StyleDaud, Noshin, Valerie Currie, Gary Duncan, Freda Farquharson, Tomoya Yoshinari, Petra Louis, and Silvia W. Gratz. 2020. "Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins" Toxins 12, no. 10: 654. https://doi.org/10.3390/toxins12100654
APA StyleDaud, N., Currie, V., Duncan, G., Farquharson, F., Yoshinari, T., Louis, P., & Gratz, S. W. (2020). Prevalent Human Gut Bacteria Hydrolyse and Metabolise Important Food-Derived Mycotoxins and Masked Mycotoxins. Toxins, 12(10), 654. https://doi.org/10.3390/toxins12100654