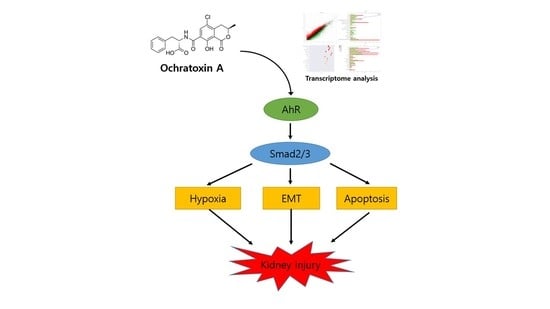
Transcriptome Analysis Reveals the AhR, Smad2/3, and HIF-1α Pathways as the Mechanism of Ochratoxin A Toxicity in Kidney Cells
Abstract
:1. Introduction
2. Results
2.1. Analysis of DEGs
2.2. Functional Annotation of DEGs
2.3. Gene Set Enrichment Analysis of Genes Related to OTA
2.4. Validation of Gene Expression Patterns Using qRT-PCR
2.5. Effects of AhR Knockdown on EMT and Kidney Injury-Related Marker Expression
2.6. Effect of AhR Knockdown on Smad2/3 and HIF-1α Expression
2.7. Effects of Smad2/3 and HIF-1α Knockdown on EMT and Renal Injury-Related Markers Expression
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. OTA Treatment of HK-2 Cells
4.4. Cytotoxicity Assay
4.5. Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.6. Isolation of Total Cell Lysate and Western Blot Analysis
4.7. Transfection with Small Interfering RNA (siRNA)
4.8. RNA-sequencing
4.9. RNA-seq Data Analysis
4.10. Statistical Analysis of Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, Z.; Huang, K.; Luo, Y. Ochratoxin A and ochratoxin-producing fungi on cereal grain in China: A review. Food Addit. Contam. Part A 2015, 32, 461–470. [Google Scholar]
- Bui-Klimke, T.R.; Wu, F. Ochratoxin A and human health risk: A review of the evidence. Crit. Rev. Food Sci. Nutr. 2015, 55, 1860–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Merwe, K.; Steyn, P.; Fourie, L.; Scott, D.B.; Theron, J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 1965, 205, 1112–1113. [Google Scholar] [CrossRef] [PubMed]
- Delacruz, L.; Bach, P. The role of ochratoxin A metabolism and biochemistry in animal and human nephrotoxicity. J. Biopharm. Sci. 1990, 1, 277–304. [Google Scholar]
- Galtier, P.; Alvinerie, M.; Charpenteau, J. The pharmacokinetic profiles of ochratoxin A in pigs, rabbits and chickens. Food Cosmet. Toxicol. 1981, 19, 735–738. [Google Scholar] [CrossRef]
- IARC. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans; World Health Organization: Geneva, Switzerland, 1993; Volume 56. [Google Scholar]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef] [PubMed]
- Mally, A.; Dekant, W. Mycotoxins and the kidney: Modes of action for renal tumor formation by ochratoxin A in rodents. Mol. Nutr. Food Res. 2009, 53, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Gan, F.; Hou, L.; Zhou, Y.; Liu, Y.; Huang, D.; Chen, X.; Huang, K. Effects of ochratoxin A on ER stress, MAPK signaling pathway and autophagy of kidney and spleen in pigs. Environ. Toxicol. 2017, 32, 2277–2286. [Google Scholar] [CrossRef]
- Damiano, S.; Iovane, V.; Squillacioti, C.; Mirabella, N.; Prisco, F.; Ariano, A.; Amenta, M.; Giordano, A.; Florio, S.; Ciarcia, R. Red orange and lemon extract prevents the renal toxicity induced by ochratoxin A in rats. J. Cell. Physiol. 2020, 235, 5386–5393. [Google Scholar] [CrossRef]
- Damiano, S.; Longobardi, C.; Andretta, E.; Prisco, F.; Piegari, G.; Squillacioti, C.; Montagnaro, S.; Pagnini, F.; Badino, P.; Florio, S. Antioxidative Effects of Curcumin on the Hepatotoxicity Induced by Ochratoxin A in Rats. Antioxidants 2021, 10, 125. [Google Scholar] [CrossRef]
- Cui, G.; Li, L.; Xu, W.; Wang, M.; Jiao, D.; Yao, B.; Xu, K.; Chen, Y.; Yang, S.; Long, M. Astaxanthin protects ochratoxin A-induced oxidative stress and apoptosis in the heart via the Nrf2 pathway. Oxidative Med. Cell. Longev. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Chen, W.; Yu, X.-Q. Long non-coding RNAs as novel diagnostic and therapeutic targets in kidney disease. Chronic Dis. Transl. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Shi, Y.; Diamond-Stanic, M.; Sharma, K. Role of Mitochondria in the Regulation of Kidney Function and Metabolism in Type 2 Diabetes. In Mitochondria in Obesity and Type 2 Diabetes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 287–300. [Google Scholar]
- Radford, R.; Frain, H.; Ryan, M.P.; Slattery, C.; McMorrow, T. Mechanisms of chemical carcinogenesis in the kidneys. Int. J. Mol. Sci. 2013, 14, 19416–19433. [Google Scholar] [CrossRef] [Green Version]
- Zingerle, M.; Silbernagl, S.; Gekle, M. Reabsorption of the nephrotoxin ochratoxin A along the rat nephron in vivo. J. Pharmacol. Exp. Ther. 1997, 280, 220–224. [Google Scholar]
- Schwerdt, G.; Bauer, K.; Gekle, M.; Silbernagl, S. Accumulation of ochratoxin A in rat kidney in vivo and in cultivated renal epithelial cells in vitro. Toxicology 1996, 114, 177–185. [Google Scholar] [CrossRef]
- Kashida, Y.; Takahashi, A.; Moto, M.; Okamura, M.; Muguruma, M.; Jin, M.; Arai, K.; Mitsumori, K. Gene expression analysis in mice liver on hepatocarcinogenesis by flumequine. Arch. Toxicol. 2006, 80, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lu, J.; Zhao, D. Toxicity and transcriptome sequencing (RNA-seq) analyses of adult zebrafish in response to exposure carboxymethyl cellulose stabilized iron sulfide nanoparticles. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, M.; Borlak, J.; Tong, W. A decade of toxicogenomic research and its contribution to toxicological science. Toxicol. Sci. 2012, 130, 217–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wentzel, J.F.; Lombard, M.J.; Du Plessis, L.H.; Zandberg, L. Evaluation of the cytotoxic properties, gene expression profiles and secondary signalling responses of cultured cells exposed to fumonisin B1, deoxynivalenol and zearalenone mycotoxins. Arch. Toxicol. 2017, 91, 2265–2282. [Google Scholar] [CrossRef]
- Afshari, C.A.; Hamadeh, H.K.; Bushel, P.R. The evolution of bioinformatics in toxicology: Advancing toxicogenomics. Toxicol. Sci. 2011, 120, S225–S237. [Google Scholar] [CrossRef] [Green Version]
- Arbillaga, L.; Azqueta, A.; van Delft, J.H.; de Cerain, A.L. In vitro gene expression data supporting a DNA non-reactive genotoxic mechanism for ochratoxin A. Toxicol. Appl. Pharmacol. 2007, 220, 216–224. [Google Scholar] [CrossRef]
- Hibi, D.; Kijima, A.; Kuroda, K.; Suzuki, Y.; Ishii, Y.; Jin, M.; Nakajima, M.; Sugita-Konishi, Y.; Yanai, T.; Nohmi, T. Molecular mechanisms underlying ochratoxin A-induced genotoxicity: Global gene expression analysis suggests induction of DNA double-strand breaks and cell cycle progression. J. Toxicol. Sci. 2013, 38, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hundhausen, C.; Boesch-Saadatmandi, C.; Matzner, N.; Lang, F.; Blank, R.; Wolffram, S.; Blaschek, W.; Rimbach, G. Ochratoxin a lowers mRNA levels of genes encoding for key proteins of liver cell metabolism. Cancer Genom. Proteom. 2008, 5, 319–332. [Google Scholar]
- Raghubeer, S.; Nagiah, S.; Chuturgoon, A.A. Acute Ochratoxin A exposure induces inflammation and apoptosis in human embryonic kidney (HEK293) cells. Toxicon 2017, 137, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mao, X.; Liu, K.; Sun, J.; Li, B.; Malyar, R.M.; Liu, D.; Pan, C.; Gan, F.; Liu, Y. Ochratoxin A induces nephrotoxicity in vitro and in vivo via pyroptosis. Arch. Toxicol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Tang, Y.-L.; Liang, X.-H. EMT: A new vision of hypoxia promoting cancer progression. Cancer Biol. Ther. 2011, 11, 714–723. [Google Scholar] [CrossRef] [Green Version]
- Myszczyszyn, A.; Czarnecka, A.M.; Matak, D.; Szymanski, L.; Lian, F.; Kornakiewicz, A.; Bartnik, E.; Kukwa, W.; Kieda, C.; Szczylik, C. The role of hypoxia and cancer stem cells in renal cell carcinoma pathogenesis. Stem Cell Rev. Rep. 2015, 11, 919–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.S.; Lee, H.J.; Pyo, M.C.; Ryu, D.; Lee, K.-W. Ochratoxin A-Induced Hepatotoxicity through Phase I and Phase II Reactions Regulated by AhR in Liver Cells. Toxins 2019, 11, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peraica, M.; Flajs, D.; Domijan, A.-M.; Ivić, D.; Cvjetković, B. Ochratoxin A contamination of food from Croatia. Toxins 2010, 2, 2098–2105. [Google Scholar] [CrossRef] [Green Version]
- Sergent, T.; Garsou, S.; Schaut, A.; De Saeger, S.; Pussemier, L.; Van Peteghem, C.; Larondelle, Y.; Schneider, Y.-J. Differential modulation of ochratoxin A absorption across Caco-2 cells by dietary polyphenols, used at realistic intestinal concentrations. Toxicol. Lett. 2005, 159, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, W.; Huang, K.; Zhang, B.; Wang, H.; Zhang, X.; Gong, L.; Luo, Y.; He, X. Precision toxicology shows that troxerutin alleviates ochratoxin A–induced renal lipotoxicity. FASEB J. 2019, 33, 2212–2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Yu, T.; Qi, X.; Gao, J.; Huang, K.; He, X.; Luo, H.; Xu, W. Limited link between oxidative stress and ochratoxin A—Induced renal injury in an acute toxicity rat model. Toxins 2016, 8, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.J.; Pyo, M.C.; Shin, H.S.; Ryu, D.; Lee, K.-W. Renal toxicity through AhR, PXR, and Nrf2 signaling pathway activation of ochratoxin A-induced oxidative stress in kidney cells. Food Chem. Toxicol. 2018, 122, 59–68. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Li, R.; Michal, J.J.; Zhang, S.; Dodson, M.V.; Zhang, Z.; Harland, R.M. Whole transcriptome analysis with sequencing: Methods, challenges and potential solutions. Cell. Mol. Life Sci. 2015, 72, 3425–3439. [Google Scholar] [CrossRef] [PubMed]
- Raghubeer, S.; Nagiah, S.; Chuturgoon, A. Ochratoxin A upregulates biomarkers associated with hypoxia and transformation in human kidney cells. Toxicol. In Vitro 2019, 57, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Schwerdt, G.; Holzinger, H.; Sauvant, C.; Königs, M.; Humpf, H.-U.; Gekle, M. Long-term effects of ochratoxin A on fibrosis and cell death in human proximal tubule or fibroblast cells in primary culture. Toxicology 2007, 232, 57–67. [Google Scholar] [CrossRef]
- Hodges, R.E.; Minich, D.M. Modulation of metabolic detoxification pathways using foods and food-derived components: A scientific review with clinical application. J. Nutr. Metab. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cederbaum, A.I. CYP2E1 potentiation of LPS and TNFα-induced hepatotoxicity by mechanisms involving enhanced oxidative and nitrosative stress, activation of MAP kinases, and mitochondrial dysfunction. Genes Nutr. 2010, 5, 149–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Gómez, G.; Rocha-Zavaleta, L.; Rodríguez-Sosa, M.; Petrosyan, P.; Rubio-Lightbourn, J. Benzo [a] pyrene activates an AhR/Src/ERK axis that contributes to CYP1A1 induction and stable DNA adducts formation in lung cells. Toxicol. Lett. 2018, 289, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W. Aryl hydrocarbon receptor (AHR):“pioneer member” of the basic-helix/loop/helix per-Arnt-sim (bHLH/PAS) family of “sensors” of foreign and endogenous signals. Prog. Lipid Res. 2017, 67, 38–57. [Google Scholar] [CrossRef] [PubMed]
- Phillips, T.D.; Richardson, M.; Cheng, Y.-S.L.; He, L.; McDonald, T.J.; Cizmas, L.H.; Safe, S.H.; Donnelly, K.C.; Wang, F.; Moorthy, B. Mechanistic relationships between hepatic genotoxicity and carcinogenicity in male B6C3F1 mice treated with polycyclic aromatic hydrocarbon mixtures. Arch. Toxicol. 2015, 89, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yang, M.; Meng, T.; Niu, Y.; Dai, Y.; Zhang, L.; Zheng, X.; Jalava, P.; Dong, G.; Gao, W. Oxidative stress induced by ultrafine carbon black particles can elicit apoptosis in vivo and vitro. Sci. Total Environ. 2020, 709, 135802. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, H.; He, H.; Ma, C. Assays for alkaline phosphatase activity: Progress and prospects. Trac Trends Anal. Chem. 2019. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Petrik, J.; Žanić-Grubišić, T.; Barišić, K.; Pepeljnjak, S.; Radić, B.; Ferenčić, Ž.; Čepelak, I. Apoptosis and oxidative stress induced by ochratoxin A in rat kidney. Arch. Toxicol. 2003, 77, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Ramyaa, P.; Padma, V.V. Ochratoxin-induced toxicity, oxidative stress and apoptosis ameliorated by quercetin–Modulation by Nrf2. Food Chem. Toxicol. 2013, 62, 205–216. [Google Scholar] [CrossRef]
- Bartis, D.; Mise, N.; Mahida, R.Y.; Eickelberg, O.; Thickett, D.R. Epithelial–mesenchymal transition in lung development and disease: Does it exist and is it important? Thorax 2014, 69, 760–765. [Google Scholar] [CrossRef] [Green Version]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Benzoubir, N.; Mussini, C.; Lejamtel, C.; Dos Santos, A.; Guillaume, C.; Desterke, C.; Samuel, D.; Brechot, C.; Bourgeade, M.-F.; Guettier, C. Gamma-smooth muscle actin expression is associated with epithelial-mesenchymal transition and stem-like properties in hepatocellular carcinoma. PLoS ONE 2015, 10, e0130559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Bu, Y.; Liu, X.; Wang, X.; Zhang, G.; Wang, E.; Ding, S.; Liu, Y.; Shi, R.; Li, Q. TCDD promoted EMT of hFPECs via AhR, which involved the activation of EGFR/ERK signaling. Toxicol. Appl. Pharmacol. 2016, 298, 48–55. [Google Scholar] [CrossRef]
- Wu, Y.; Niu, Y.; Leng, J.; Xu, J.; Chen, H.; Li, H.; Wang, L.; Hu, J.; Xia, D.; Wu, Y. Benzo (a) pyrene regulated A549 cell migration, invasion and epithelial-mesenchymal transition by up-regulating long non-coding RNA linc00673. Toxicol. Lett. 2020, 320, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Inazaki, K.; Kanamaru, Y.; Kojima, Y.; Sueyoshi, N.; Okumura, K.; Kaneko, K.; Yamashiro, Y.; Ogawa, H.; Nakao, A. Smad3 deficiency attenuates renal fibrosis, inflammation, and apoptosis after unilateral ureteral obstruction. Kidney Int. 2004, 66, 597–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, D.; Lyman, S.; Qian, T.; Lemasters, J.J.; Rippe, R.A.; Nitta, T.; Kim, J.-S.; Behrns, K.E. Transforming growth factor beta mediates hepatocyte apoptosis through Smad3 generation of reactive oxygen species. Biochimie 2007, 89, 1464–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, M.; Tanaka, T.; Yamamoto, T.; Noiri, E.; Miyata, T.; Inagi, R.; Fujita, T.; Nangaku, M. Hypoperfusion of peritubular capillaries induces chronic hypoxia before progression of tubulointerstitial injury in a progressive model of rat glomerulonephritis. J. Am. Soc. Nephrol. 2004, 15, 1574–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, Y.; Tanaka, T.; Nangaku, M. Renal hypoxia in CKD; pathophysiology and detecting methods. Front. Physiol. 2017, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Colgan, S.P.; Shelley, C.S. Hypoxia: The force that drives chronic kidney disease. Clin. Med. Res. 2016, 14, 15–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erler, J.T.; Cawthorne, C.J.; Williams, K.J.; Koritzinsky, M.; Wouters, B.G.; Wilson, C.; Miller, C.; Demonacos, C.; Stratford, I.J.; Dive, C. Hypoxia-mediated down-regulation of Bid and Bax in tumors occurs via hypoxia-inducible factor 1-dependent and-independent mechanisms and contributes to drug resistance. Mol. Cell. Biol. 2004, 24, 2875–2889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhao, S.; Nakada, K.; Kuge, Y.; Tamaki, N.; Okada, F.; Wang, J.; Shindo, M.; Higashino, F.; Takeda, K. Dominant-negative hypoxia-inducible factor-1α reduces tumorigenicity of pancreatic cancer cells through the suppression of glucose metabolism. Am. J. Pathol. 2003, 162, 1283–1291. [Google Scholar] [CrossRef]
- Dai, S.; Huang, M.L.; Hsu, C.Y.; Chao, K.C. Inhibition of hypoxia inducible factor 1α causes oxygen-independent cytotoxicity and induces p53 independent apoptosis in glioblastoma cells. Int. J. Radiat. Oncol. Biol. Phys. 2003, 55, 1027–1036. [Google Scholar] [CrossRef]
- Kang, D.-H.; Hughes, J.; Mazzali, M.; Schreiner, G.F.; Johnson, R.J. Impaired angiogenesis in the remnant kidney model: II. Vascular endothelial growth factor administration reduces renal fibrosis and stabilizes renal function. J. Am. Soc. Nephrol. 2001, 12, 1448–1457. [Google Scholar] [PubMed]
- Agarwal, A.; Nick, H.S. Renal response to tissue injury: Lessons from heme oxygenase-1 gene ablation and expression. J. Am. Soc. Nephrol. 2000, 11, 965–973. [Google Scholar] [PubMed]
- Vesey, D.A.; Cheung, C.; Pat, B.; Endre, Z.; Gobe, G.; Johnson, D.W. Erythropoietin protects against ischaemic acute renal injury. Nephrol. Dial. Transplant. 2004, 19, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Conde, E.; Alegre, L.; Blanco-Sanchez, I.; Saenz-Morales, D.; Aguado-Fraile, E.; Ponte, B.; Ramos, E.; Sáiz, A.; Jiménez, C.; Ordoñez, A. Hypoxia inducible factor 1-alpha (HIF-1 alpha) is induced during reperfusion after renal ischemia and is critical for proximal tubule cell survival. PLoS ONE 2012, 7, e33258. [Google Scholar] [CrossRef]
- Hill, P.; Shukla, D.; Tran, M.G.; Aragones, J.; Cook, H.T.; Carmeliet, P.; Maxwell, P.H. Inhibition of hypoxia inducible factor hydroxylases protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2008, 19, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danobeitia, J.S.; Djamali, A.; Fernandez, L.A. The role of complement in the pathogenesis of renal ischemia-reperfusion injury and fibrosis. Fibrogenesis Tissue Repair 2014, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kapitsinou, P.P.; Jaffe, J.; Michael, M.; Swan, C.E.; Duffy, K.J.; Erickson-Miller, C.L.; Haase, V.H. Preischemic targeting of HIF prolyl hydroxylation inhibits fibrosis associated with acute kidney injury. Am. J. Physiol. Ren. Physiol. 2012, 302, F1172–F1179. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-Y.; Pyo, M.C.; Nam, M.-H.; Lee, K.-W. ERK/Nrf2 pathway activation by caffeic acid in HepG2 cells alleviates its hepatocellular damage caused by t-butylhydroperoxide-induced oxidative stress. BMC Complementary Altern. Med. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Turner, A.; Aggarwal, P.; Matter, A.; Storvick, E.; Arnett, D.K.; Broeckel, U. Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]








| Gene Set Name | # Genes in Gene Set (K) | # Genes in Overlap (k) | k/K | p-Value | FDR
q-Value |
|---|---|---|---|---|---|
| HYPOXIA | 200 | 57 | 0.285 | 1.02 × 10−27 | 5.09 × 10−26 |
| TNFA_SIGNALING_VIA_NFKB | 200 | 50 | 0.250 | 1.05 × 10−21 | 2.62 × 10−20 |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 200 | 46 | 0.230 | 1.60 × 10−18 | 2.66 × 10−17 |
| GLYCOLYSIS | 200 | 44 | 0.220 | 5.26 × 10−17 | 6.58 × 10−16 |
| MTORC1_SIGNALING | 200 | 43 | 0.215 | 2.89 × 10−16 | 2.89 × 10−15 |
| UV_RESPONSE_DN | 144 | 35 | 0.243 | 3.19 × 10−15 | 2.65 × 10−14 |
| OXIDATIVE_PHOSPHORYLATION | 200 | 40 | 0.200 | 4.02 × 10−14 | 2.87 × 10−13 |
| UNFOLDED_PROTEIN_RESPONSE | 113 | 29 | 0.257 | 1.63 × 10−13 | 1.02 × 10−12 |
| ADIPOGENESIS | 200 | 39 | 0.195 | 1.96 × 10−13 | 1.09 × 10−12 |
| MITOTIC_SPINDLE | 199 | 38 | 0.191 | 7.86 × 10−13 | 3.85 × 10−12 |
| INFLAMMATORY_RESPONSE | 200 | 38 | 0.190 | 9.25 × 10−13 | 3.85 × 10−12 |
| P53_PATHWAY | 200 | 38 | 0.190 | 9.25 × 10−13 | 3.85 × 10−12 |
| INTERFERON_GAMMA_RESPONSE | 200 | 35 | 0.175 | 7.99 × 10−11 | 3.07 × 10−10 |
| ESTROGEN_RESPONSE_LATE | 200 | 34 | 0.170 | 3.30 × 10−10 | 1.18 × 10−9 |
| APOPTOSIS | 161 | 30 | 0.186 | 3.65 × 10−10 | 1.22 × 10−9 |
| TGF_BETA_SIGNALING | 54 | 17 | 0.315 | 5.17 × 10−10 | 1.61 × 10−9 |
| ESTROGEN_RESPONSE_EARLY | 200 | 30 | 0.150 | 6.75 × 10−8 | 1.88 × 10−7 |
| IL2_STAT5_SIGNALING | 200 | 30 | 0.150 | 6.75 × 10−8 | 1.88 × 10−7 |
| COAGULATION | 138 | 24 | 0.174 | 8.07 × 10−8 | 2.12 × 10−7 |
| XENOBIOTIC_METABOLISM | 200 | 32 | 0.160 | 1.86 × 10−8 | 4.05 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyo, M.C.; Choi, I.-G.; Lee, K.-W. Transcriptome Analysis Reveals the AhR, Smad2/3, and HIF-1α Pathways as the Mechanism of Ochratoxin A Toxicity in Kidney Cells. Toxins 2021, 13, 190. https://doi.org/10.3390/toxins13030190
Pyo MC, Choi I-G, Lee K-W. Transcriptome Analysis Reveals the AhR, Smad2/3, and HIF-1α Pathways as the Mechanism of Ochratoxin A Toxicity in Kidney Cells. Toxins. 2021; 13(3):190. https://doi.org/10.3390/toxins13030190
Chicago/Turabian StylePyo, Min Cheol, In-Geol Choi, and Kwang-Won Lee. 2021. "Transcriptome Analysis Reveals the AhR, Smad2/3, and HIF-1α Pathways as the Mechanism of Ochratoxin A Toxicity in Kidney Cells" Toxins 13, no. 3: 190. https://doi.org/10.3390/toxins13030190
APA StylePyo, M. C., Choi, I. -G., & Lee, K. -W. (2021). Transcriptome Analysis Reveals the AhR, Smad2/3, and HIF-1α Pathways as the Mechanism of Ochratoxin A Toxicity in Kidney Cells. Toxins, 13(3), 190. https://doi.org/10.3390/toxins13030190