Controlled Production of Zearalenone-Glucopyranoside Standards with Cunninghamella Strains Using Sulphate-Depleted Media
Abstract
:1. Introduction
2. Results and Discussion
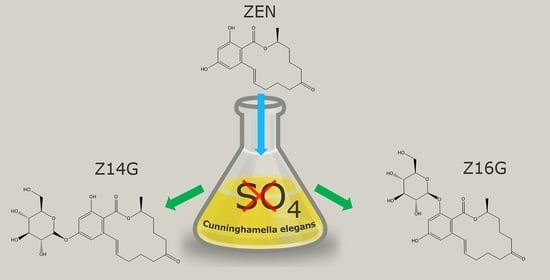
2.1. Small-Scale Biotransformation of ZEN
2.2. Upscaled Production of ZEN-Glycosides
2.3. NMR Analysis of Purified Z14G and Z16G Fractions
2.4. Feasibility of Biotransformation for Other ZEN Metabolites
2.5. Feasibility of Biotransformation for Entirely Different Mycotoxins
3. Conclusions
4. Materials and Methods
4.1. Instrumentation
4.2. Materials
4.3. Fungal Starter Cultures
4.4. Biotransformation of ZEN in Liquid Fungal Cultures
4.5. LC-MS/MS Analysis of Produced Conjugates
4.6. NMR Analysis of Produced Conjugates
4.7. LC-HRMS Analysis of the ZEN Metabolites by Cunninghamella and Collection of HRMS Spectra of the Purified ZEN Glycosides
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zinedine, A.; Soriano, J.M.; Moltó, J.C.; Mañes, J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: An oestrogenic mycotoxin. Food Chem. Toxicol. 2007, 45, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chain, E.P.O.C.I.T.F. Scientific Opinion on the risks for public health related to the presence of zearalenone in food. EFSA J. 2011, 9, 2197. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain (CONTAM). Appropriateness to set a group health-based guidance value for zearalenone and its modified forms. EFSA J. 2016, 14, 4425. [Google Scholar] [CrossRef]
- Golinski, P.; Waskiewicz, A.; Gromadzka, K. Zearalenone and its Derivatives: Known Toxins in New Aspects. In Mycotoxins in Food, Feed and Bioweapons; Rai, M., Varma, A., Eds.; Springer: Berlin, Germany, 2010; pp. 113–129. [Google Scholar]
- Metzler, M. Proposal for a uniform designation of zearalenone and its metabolites. Mycotoxin Res. 2010, 27, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Nie, D.; Fan, K.; Yang, J.; Guo, W.; Meng, J.; Zhao, Z.; Han, Z. A systematic review of plant-conjugated masked mycotoxins: Occurrence, toxicology, and metabolism. Crit. Rev. Food Sci. Nutr. 2019, 60, 1523–1537. [Google Scholar] [CrossRef]
- Berthiller, F.; Schuhmacher, R.; Adam, G.; Krska, R. Formation, determination and significance of masked and other conjugated mycotoxins. Anal. Bioanal. Chem. 2009, 395, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; De Saeger, S.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2012, 57, 165–186. [Google Scholar] [CrossRef]
- Schneweis, I.; Meyer, K.; Engelhardt, G.; Bauer, J. Occurrence of Zearalenone-4-β-d-glucopyranoside in Wheat. J. Agric. Food Chem. 2002, 50, 1736–1738. [Google Scholar] [CrossRef]
- De Boevre, M.; Jacxsens, L.; Lachat, C.; Eeckhout, M.; Di Mavungu, J.D.; Audenaert, K.; Maene, P.; Haesaert, G.; Kolsteren, P.; De Meulenaer, B.; et al. Human exposure to mycotoxins and their masked forms through cereal-based foods in Belgium. Toxicol. Lett. 2013, 218, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Streit, E.; Schwab, C.; Sulyok, M.; Naehrer, K.; Krska, R.; Schatzmayr, G. Multi-Mycotoxin Screening Reveals the Occurrence of 139 Different Secondary Metabolites in Feed and Feed Ingredients. Toxins 2013, 5, 504–523. [Google Scholar] [CrossRef] [Green Version]
- Nathanail, A.V.; Syvähuoko, J.; Malachová, A.; Jestoi, M.; Varga, E.; Michlmayr, H.; Adam, G.; Sieviläinen, E.; Berthiller, F.; Peltonen, K. Simultaneous determination of major type A and B trichothecenes, zearalenone and certain modified metabolites in Finnish cereal grains with a novel liquid chromatography-tandem mass spectrometric method. Anal. Bioanal. Chem. 2015, 407, 4745–4755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.; Van Dam, R.; Van Doorn, R.; Katerere, D.; Berthiller, F.; Haasnoot, W.; Nielen, M.W.F. Mycotoxin profiling of 1000 beer samples with a special focus on craft beer. PLoS ONE 2017, 12, e0185887. [Google Scholar] [CrossRef] [Green Version]
- Borzekowski, A.; Anggriawan, R.; Auliyati, M.; Kunte, H.-J.; Koch, M.; Rohn, S.; Karlovsky, P.; Maul, R. Formation of Zearalenone Metabolites in Tempeh Fermentation. Molecules 2019, 24, 2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gareis, M.; Bauer, J.; Thiem, J.; Plank, G.; Grabley, S.; Gedek, B. Cleavage of Zearalenone-Glycoside, a “Masked” Mycotoxin, during Digestion in Swine. J. Veter. Med. Ser. B 1990, 37, 236–240. [Google Scholar] [CrossRef]
- Binder, S.B.; Schwartz-Zimmermann, H.E.; Varga, E.; Bichl, G.; Michlmayr, H.; Adam, G.; Berthiller, F. Metabolism of Zearalenone and Its Major Modified Forms in Pigs. Toxins 2017, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catteuw, A.; Broekaert, N.; De Baere, S.; Lauwers, M.; Gasthuys, E.; Huybrechts, B.; Callebaut, A.; Ivanova, L.; Uhlig, S.; De Boevre, M.; et al. Insights into In Vivo Absolute Oral Bioavailability, Biotransformation, and Toxicokinetics of Zearalenone, α-Zearalenol, β-Zearalenol, Zearalenone-14-glucoside, and Zearalenone-14-sulfate in Pigs. J. Agric. Food Chem. 2019, 67, 3448–3458. [Google Scholar] [CrossRef]
- Dellafiora, L.; Galaverna, G.; Righi, F.; Cozzini, P.; Dall’Asta, C. Assessing the hydrolytic fate of the masked mycotoxin zearalenone-14-glucoside—A warning light for the need to look at the “maskedome”. Food Chem. Toxicol. 2017, 99, 9–16. [Google Scholar] [CrossRef]
- Veršilovskis, A.; Geys, J.; Huybrechts, B.; Goossens, E.; De Saeger, S.; Callebaut, A. Simultaneous determination of masked forms of deoxynivalenol and zearalenone after oral dosing in rats by LC-MS/MS. World Mycotoxin J. 2012, 5, 303–318. [Google Scholar] [CrossRef]
- Paris, M.P.K.; Schweiger, W.; Hametner, C.; Stückler, R.; Muehlbauer, G.J.; Varga, E.; Krska, R.; Berthiller, F.; Adam, G. Zearalenone-16-O-glucoside: A New Masked Mycotoxin. J. Agric. Food Chem. 2014, 62, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Werner, U.; Sulyok, M.; Krska, R.; Hauser, M.-T.; Schuhmacher, R. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) determination of phase II metabolites of the mycotoxin zearalenone in the model plantArabidopsis thaliana. Food Addit. Contam. 2006, 23, 1194–1200. [Google Scholar] [CrossRef] [Green Version]
- Poppenberger, B.; Berthiller, F.; Bachmann, H.; Lucyshyn, D.; Peterbauer, C.; Mitterbauer, R.; Schuhmacher, R.; Krska, R.; Glössl, J.; Adam, G. Heterologous Expression of Arabidopsis UDP-Glucosyltransferases in Saccharomyces cerevisiae for Production of Zearalenone-4-O-Glucoside. Appl. Environ. Microbiol. 2006, 72, 4404–4410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthiller, F.; Hametner, C.; Krenn, P.; Schweiger, W.; Ludwig, R.; Adam, G.; Krska, R.; Schuhmacher, R. Preparation and characterization of the conjugatedFusariummycotoxins zearalenone-4O-β-D-glucopyranoside, α-zearalenol-4O-β-D-glucopyranoside and β-zearalenol-4O-β-D-glucopyranoside by MS/MS and two-dimensional NMR. Food Addit. Contam. Part A 2009, 26, 207–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righetti, L.; Rolli, E.; Galaverna, G.; Suman, M.; Bruni, R.; Dall’Asta, C. Plant organ cultures as masked mycotoxin biofactories: Deciphering the fate of zearalenone in micropropagated durum wheat roots and leaves. PLoS ONE 2017, 12, e0187247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolli, E.; Righetti, L.; Galaverna, G.; Suman, M.; Dall’Asta, C.; Bruni, R. Zearalenone Uptake and Biotransformation in MicropropagatedTriticum durumDesf. Plants: A Xenobolomic Approach. J. Agric. Food Chem. 2018, 66, 1523–1532. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Genet. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Wisecaver, J.H.; Slot, J.C.; Rokas, A. The Evolution of Fungal Metabolic Pathways. PLoS Genet. 2014, 10, e1004816. [Google Scholar] [CrossRef] [Green Version]
- Hegazy, M.-E.F.; Mohamed, T.A.; ElShamy, A.I.; Mohamed, A.-E.-H.H.; Mahalel, U.A.; Reda, E.H.; Shaheen, A.M.; Tawfik, W.A.; Shahat, A.A.; Shams, K.A.; et al. Microbial biotransformation as a tool for drug development based on natural products from mevalonic acid pathway: A review. J. Adv. Res. 2015, 6, 17–33. [Google Scholar] [CrossRef] [Green Version]
- Asha, S.; Vidyavathi, M. Cunninghamella—A microbial model for drug metabolism studies—A review. Biotechnol. Adv. 2009, 27, 16–29. [Google Scholar] [CrossRef]
- Li, P.; Su, R.; Yin, R.; Lai, D.; Wang, M.; Liu, Y.; Zhou, L. Detoxification of Mycotoxins through Biotransformation. Toxins 2020, 12, 121. [Google Scholar] [CrossRef] [Green Version]
- Brodehl, A.; Möller, A.; Kunte, H.-J.; Koch, M.; Maul, R. Biotransformation of the mycotoxin zearalenone by fungi of the generaRhizopusandAspergillus. FEMS Microbiol. Lett. 2014, 359, 124–130. [Google Scholar] [CrossRef] [Green Version]
- Borzekowski, A.; Drewitz, T.; Keller, J.; Pfeifer, D.; Kunte, H.-J.; Koch, M.; Rohn, S.; Maul, R. Biosynthesis and Characterization of Zearalenone-14-Sulfate, Zearalenone-14-Glucoside and Zearalenone-16-Glucoside Using Common Fungal Strains. Toxins 2018, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- El-Sharkawy, S.; Abul-Hajj, Y. Microbial Transformation of Zearalenone, I. Formation of Zearalenone-4-O-β-glucoside. J. Nat. Prod. 1987, 50, 520–521. [Google Scholar] [CrossRef]
- El-Sharkaway, S.H.; Selim, M.I.; Afifi, M.S.; Halaweish, F.T. Microbial transformation of zearalenone to a zearalenone sulfate. Appl. Environ. Microbiol. 1991, 57, 549–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zi, J.; Valiente, J.; Zeng, J.; Zhan, J. Metabolism of quercetin by Cunninghamella elegans ATCC 9245. J. Biosci. Bioeng. 2011, 112, 360–362. [Google Scholar] [CrossRef]
- Orabi, K.Y.; Li, E.; Clark, A.M.; Hufford, C.D. Microbial Transformation of Sampangine. J. Nat. Prod. 1999, 62, 988–992. [Google Scholar] [CrossRef]
- Rychlik, M.; Humpf, H.-U.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [CrossRef] [Green Version]
- McCormick, S.P.; Price, N.P.J.; Kurtzman, C.P. Glucosylation and Other Biotransformations of T-2 Toxin by Yeasts of the Trichomonascus Clade. Appl. Environ. Microbiol. 2012, 78, 8694–8702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, J.; Péteri, Z.; Tábori, K.; Téren, J.; Vágvölgyi, C. Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 2005, 99, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Ruhland, M.; Engelhardt, G.; Wallnöfer, P.R. Transformation of the mycotoxin ochratoxin A in plants. Time course and rates of degradation and metabolite production in cell-suspension cultures of different crop plants. Mycopathologia 1996, 134, 97–102. [Google Scholar] [CrossRef]
- Ruhland, M.; Engelhardt, G. Transformation of the mycotoxin ochratoxin A in wheat and maize cell suspension cultures. Naturwissenschaften 1994, 81, 453–454. [Google Scholar] [CrossRef]
- Mohamed, S.S.; Shafei, M.S.; Allam, R.; Elazzazy, A.; Abo Elsoud, M.; El-Menoufy, H.A. Effect of aeration rate on the biotransformation of cortexolone using Cunninghamella elegans in a laboratory scale bioreactor. World Appl. Sci. J. 2013, 25, 176–183. [Google Scholar] [CrossRef]





| Potato Dextrose Broth Medium (PDB) | Sulphate-Depleted Medium (MCD) | ||||||
|---|---|---|---|---|---|---|---|
| Compound | MW | Amount (µg) | Amount (µM) | Conversion (%) | Amount (µg) | Amount (µM) | Conversion (%) |
| ZEN | 318.4 | 2000 | 6.28 | n.d. | 2000 | 6.28 | n.d. |
| Z14G | 480.1 | 80 | 0.167 | 3 | 1960 | 4.08 | 65 |
| Z16G | 480.1 | 56 | 0.117 | 2 | 440 | 0.92 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peters, J.; Ash, E.; Gerssen, A.; Van Dam, R.; Franssen, M.C.R.; Nielen, M.W.F. Controlled Production of Zearalenone-Glucopyranoside Standards with Cunninghamella Strains Using Sulphate-Depleted Media. Toxins 2021, 13, 366. https://doi.org/10.3390/toxins13060366
Peters J, Ash E, Gerssen A, Van Dam R, Franssen MCR, Nielen MWF. Controlled Production of Zearalenone-Glucopyranoside Standards with Cunninghamella Strains Using Sulphate-Depleted Media. Toxins. 2021; 13(6):366. https://doi.org/10.3390/toxins13060366
Chicago/Turabian StylePeters, Jeroen, Edward Ash, Arjen Gerssen, Ruud Van Dam, Maurice C. R. Franssen, and Michel W. F. Nielen. 2021. "Controlled Production of Zearalenone-Glucopyranoside Standards with Cunninghamella Strains Using Sulphate-Depleted Media" Toxins 13, no. 6: 366. https://doi.org/10.3390/toxins13060366
APA StylePeters, J., Ash, E., Gerssen, A., Van Dam, R., Franssen, M. C. R., & Nielen, M. W. F. (2021). Controlled Production of Zearalenone-Glucopyranoside Standards with Cunninghamella Strains Using Sulphate-Depleted Media. Toxins, 13(6), 366. https://doi.org/10.3390/toxins13060366