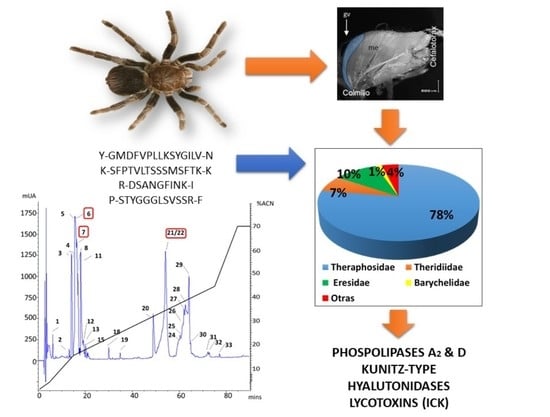
Analysis of High Molecular Mass Compounds from the Spider Pamphobeteus verdolaga Venom Gland. A Transcriptomic and MS ID Approach
Abstract
:1. Introduction
2. Results
2.1. Transcriptomic Findings
2.2. MS Findings
| rp-HPLC | Peptide Sequence | Similarity | Best Match | Protein Family |
|---|---|---|---|---|
| 6 | Y-GMDFVPLLKSYGILV-N | 100% | c51925_g1_i1 | Hyaluronidase |
| 6 | R-TIKDWYK-G | 80% | c40556_g1_i1 | Lycotoxin |
| 21–22 | K-SFPTVLTSSSMSFTK-K | 84.6% | c9919_g1_i1 | CRISP |
| R-TGPQVKGEK-S | 77.8% | c9919_g1_i1 | ||
| K-DWYKEIK-D | 55.2% | c9919_g1_i1 | ||
| K-VATGKETQYSMPK-A | 100% | c9919_g1_i1 | ||
| 7 | R-DSANGFINK-I | 73% | c14372_g1_i1 | Phospholipases D |
| K-ESGYNDK-Y | ||||
| 6 | P-STYGGGLSVSSR-F | 42.6% | c66767_g1_i1 | Kunitz-type |
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Spider Collection and Venom Extraction
5.2. Venom Fractionation
5.3. Proteomic Analysis
LC-MS/MS
5.4. Data Analysis
5.5. Transcriptomic Analysis
5.6. Nomenclature
5.7. Signal Peptide and Disulfide Bond Prediction
5.8. Data Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- King, G.F.; Hardy, M.C. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Long, R.W.; Wu, Y.Q.; Guo, Y.B.; Liu, D.L.; Peng, L.; Li, D.Q.; Yang, D.W.; Xu, X.; Liu, F.X.; et al. Identification and characterization of toxins in the venom gland of the Chinese bird spider, Haplopelma hainanum, by transcriptomic analysis. Insect Sci. 2016, 23, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Peng, L.; Chen, J.; Zhang, Y.; Xiong, X.; Liang, S. Molecular diversification based on analysis of expressed sequence tags from the venom glands of the Chinese bird spider Ornithoctonus huwena. Toxicon 2008, 51, 1479–1489. [Google Scholar] [CrossRef]
- Yuan, C.; Jin, Q.; Tang, X.; Hu, W.; Cao, R.; Yang, S.; Xiong, J.; Xie, C.; Xie, J.; Liang, S. Proteomic and peptidomic characterization of the venom from the Chinese bird spider, Ornithoctonus huwena Wang. J. Proteome Res. 2007, 6, 2792–2801. [Google Scholar] [CrossRef]
- Borges, M.H.; Figueiredo, S.G.; Leprevost, F.V.; De Lima, M.E.; Cordeiro Mdo, N.; Diniz, M.R.; Moresco, J.; Carvalho, P.C.; Yates, J.R. Venomous extract protein profile of Brazilian tarantula Grammostola iheringi: Searching for potential biotechnological applications. J. Proteom. 2016, 136, 35–47. [Google Scholar] [CrossRef]
- Liao, Z.; Cao, J.; Li, S.; Yan, X.; Hu, W.; He, Q.; Chen, J.; Tang, J.; Xie, J.; Liang, S. Proteomic and peptidomic analysis of the venom from Chinese tarantula Chilobrachys jingzhao. Proteomics 2007, 7, 1892–1907. [Google Scholar] [CrossRef]
- Cifuentes, Y.; Estrada-Gomez, S.; Vargas Munoz, L.J.; Perafan, C. Description and molecular characterization of a new species of tarantula, Pamphobeteus verdolaga, from Colombia (Aranae: Mygalomorphae: Theraphosidae). Zoologia 2016, 33. [Google Scholar] [CrossRef] [Green Version]
- Estrada-Gomez, S.; Vargas Munoz, L.J.; Quintana Castillo, J.C. Extraction and partial characterization of venom from the Colombian spider Pamphobeteus aff. nigricolor (Aranae:Theraphosidae). Toxicon 2013, 76C, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Gomez, S.; Vargas-Munoz, L.J.; Saldarriaga-Cordoba, M.; Cifuentes, Y.; Perafan, C. Identifying different transcribed proteins in the newly described Theraphosidae Pamphobeteus verdolaga. Toxicon 2017, 129, 81–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-Gomez, S.; Caldas Cardoso, F.; Vargas-Munoz, L.J.; Quintana-Castillo, J.C.; Arenas Gomez, C.M.; Pineda, S.S. Venomic, transcriptomic, and bioactivity analyses of Pamphobeteus verdolaga venom reveal complex disulfide-rich peptides that modulate calcium channels. Toxins 2019, 11, 496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clement, H.; Olvera, A.; Rodriguez, M.; Zamudio, F.; Palomares, L.A.; Possani, L.D.; Odell, G.V.; Alagon, A.; Sanchez-Lopez, R. Identification, cDNA cloning and heterologous expression of a hyaluronidase from the tarantula Brachypelma vagans venom. Toxicon 2012, 60, 1223–1227. [Google Scholar] [CrossRef]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef]
- Vargas Munoz, L.J.; Estrada-Gomez, S. Purification and Characterization of Venom Components as Source for Antibiotics. Mini-Rev. Org. Chem. 2014, 11, 15–27. [Google Scholar] [CrossRef]
- Vargas Munoz, L.J.; Estrada-Gomez, S.; Escobar, J. Snake and scorpion toxins venoms, a natural source of molecules with antimicrobial activity. Curare 2015, 2. [Google Scholar] [CrossRef]
- Kini, R.M. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon 2005, 45, 1147–1161. [Google Scholar] [CrossRef]
- Zhu, H.; Dupureur, C.M.; Zhang, X.; Tsai, M.D. Phospholipase A2 engineering. The roles of disulfide bonds in structure, conformational stability, and catalytic function. Biochemistry 1995, 34, 15307–15314. [Google Scholar] [CrossRef] [PubMed]
- Ho, I.C.; Arm, J.P.; Bingham, C.O., 3rd; Choi, A.; Austen, K.F.; Glimcher, L.H. A novel group of phospholipase A2s preferentially expressed in type 2 helper T cells. J. Biol. Chem. 2001, 276, 18321–18326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haney, R.A.; Clarke, T.H.; Gadgil, R.; Fitzpatrick, R.; Hayashi, C.Y.; Ayoub, N.A.; Garb, J.E. Effects of Gene Duplication, Positive Selection, and Shifts in Gene Expression on the Evolution of the Venom Gland Transcriptome in Widow Spiders. Genome Biol. Evol. 2016, 8, 228–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas, A.E.; Carmo, A.O.; Horta, C.C.; Leal, H.G.; Oliveira-Mendes, B.B.; Martins, A.P.; Chavez-Olortegui, C.; Kalapothakis, E. Description of Loxtox protein family and identification of a new group of Phospholipases D from Loxosceles similis venom gland. Toxicon 2016, 120, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Moreira, D.; Souza, F.N.; Fogaça, R.T.; Mangili, O.C.; Gremski, W.; Senff-Ribeiro, A.; Chaim, O.M.; Veiga, S.S. The relationship between calcium and the metabolism of plasma membrane phospholipids in hemolysis induced by brown spider venom phospholipase-D toxin. J. Cell. Biochem. 2011, 112, 2529–2540. [Google Scholar] [CrossRef]
- Yao, Y.; Li, J.; Lin, Y.; Zhou, J.; Zhang, P.; Xu, Y. Structural insights into phospholipase D function. Prog. Lipid Res. 2021, 81, 101070. [Google Scholar] [CrossRef] [PubMed]
- Kudo, I.; Murakami, M. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 2002, 68–69, 3–58. [Google Scholar] [CrossRef]
- Earl, S.T.H.; Richards, R.; Johnson, L.A.; Flight, S.; Anderson, S.; Liao, A.; De Jersey, J.; Masci, P.P.; Lavin, M.F. Identification and characterisation of Kunitz-type plasma kallikrein inhibitors unique to Oxyuranus sp. snake venoms. Biochimie 2012, 94, 365–373. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Mackessy, S.P.; Dutta, S. Characterization of a Kunitz-type protease inhibitor peptide (Rusvikunin) purified from Daboia russelii russelii venom. Int. J. Biol. Macromol. 2014, 67, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Lee, K.S.; Choo, Y.M.; Kong, D.; Yoon, H.J.; Jin, B.R. Molecular cloning and antifibrinolytic activity of a serine protease inhibitor from bumblebee (Bombus terrestris) venom. Toxicon 2013, 63, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.-s.; Chung, C.; Huang, H.-B.; Lin, S.-r. Purification and Characterization of a Chymotrypsin Inhibitor from the Venom of Ophiophagus hannah (King Cobra). Biochem. Biophys. Res. Commun. 2001, 283, 862–867. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Lu, Z.; Zhai, L.; Jiang, J.; Liu, J.; Yu, H. A novel serine protease inhibitor from the venom of Vespa bicolor Fabricius. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2009, 153, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Lee, K.S.; Kim, B.Y.; Zou, F.M.; Yoon, H.J.; Je, Y.H.; Li, J.; Jin, B.R. A spider-derived Kunitz-type serine protease inhibitor that acts as a plasmin inhibitor and an elastase inhibitor. PLoS ONE 2013, 8, e53343. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhang, Y.; Hu, W.; Xu, D.; Tao, H.; Yang, X.; Li, Y.; Jiang, L.; Liang, S. Molecular diversification of peptide toxins from the tarantula Haplopelma hainanum (Ornithoctonus hainana) venom based on transcriptomic, peptidomic, and genomic analyses. J. Proteome Res. 2010, 9, 2550–2564. [Google Scholar] [CrossRef]
- Yuan, C.H.; He, Q.Y.; Peng, K.; Diao, J.B.; Jiang, L.P.; Tang, X.; Liang, S.P. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE 2008, 3, e3414. [Google Scholar] [CrossRef]
- Undheim, E.A.; Sunagar, K.; Herzig, V.; Kely, L.; Low, D.H.; Jackson, T.N.; Jones, A.; Kurniawan, N.; King, G.F.; Ali, S.A.; et al. A proteomics and transcriptomics investigation of the venom from the barychelid spider Trittame loki (brush-foot trapdoor). Toxins 2013, 5, 2488–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zweckstetter, M.; Czisch, M.; Mayer, U.; Chu, M.L.; Zinth, W.; Timpl, R.; Holak, T.A. Structure and multiple conformations of the kunitz-type domain from human type VI collagen alpha3(VI) chain in solution. Structure 1996, 4, 195–209. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Duan, Z.; Yu, Y.; Liu, Z.; Liu, Z.; Liang, S. The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis. PLoS ONE 2013, 8, e81357. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Yan, L.; Adams, M.E. Lycotoxins, antimicrobial peptides from venom of the wolf spider Lycosa carolinensis. J. Biol. Chem. 1998, 273, 2059–2066. [Google Scholar] [CrossRef] [Green Version]
- Hughes, S.R.; Dowd, P.F.; Hector, R.E.; Panavas, T.; Sterner, D.E.; Qureshi, N.; Bischoff, K.M.; Bang, S.S.; Mertens, J.A.; Johnson, E.T.; et al. Lycotoxin-1 insecticidal peptide optimized by amino acid scanning mutagenesis and expressed as a coproduct in an ethanologenic Saccharomyces cerevisiae strain. J. Pept. Sci. 2008, 14, 1039–1050. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, L.; Jiang, L.; Meng, E.; Zhang, Y.; Xiong, X.; Liang, S. Transcriptome analysis revealed novel possible venom components and cellular processes of the tarantula Chilobrachys jingzhao venom gland. Toxicon 2008, 52, 794–806. [Google Scholar] [CrossRef]
- Diego-Garcia, E.; Peigneur, S.; Waelkens, E.; Debaveye, S.; Tytgat, J. Venom components from Citharischius crawshayi spider (Family Theraphosidae): Exploring transcriptome, venomics, and function. Cell. Mol. Life Sci. 2010, 67, 2799–2813. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Q.; Yin, W.; Zhang, X.; Huang, Y.; Luo, Y.; Qiu, P.; Su, X.; Yu, J.; Hu, S.; et al. Transcriptome analysis of Deinagkistrodon acutus venomous gland focusing on cellular structure and functional aspects using expressed sequence tags. BMC Genom. 2006, 7, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S. Proteome and peptidome profiling of spider venoms. Expert Rev. Proteom. 2008, 5, 731–746. [Google Scholar] [CrossRef]
- Duan, Z.G.; Yan, X.J.; He, X.Z.; Zhou, H.; Chen, P.; Cao, R.; Xiong, J.X.; Hu, W.J.; Wang, X.C.; Liang, S.P. Extraction and protein component analysis of venom from the dissected venom glands of Latrodectus tredecimguttatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 145, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Gething, M.-J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Pedrosa Mde, F.; Junqueira-de-Azevedo Ide, L.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279. [Google Scholar] [CrossRef] [Green Version]
- Laedermann, C.J.; Decosterd, I.; Abriel, H. Ubiquitylation of voltage-gated sodium channels. Handb. Exp. Pharmacol. 2014, 221, 231–250. [Google Scholar] [CrossRef]
- Cui, Y.; Zhu, Y.; Lin, Y.; Chen, L.; Feng, Q.; Wang, W.; Xiang, H. New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm. BMC Genom. 2018, 19, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oukkache, N.; Chgoury, F.; Lalaoui, M.; Cano, A.A.; Ghalim, N. Comparison between two methods of scorpion venom milking in Morocco. J. Venom. Anim. Toxins Incl. Trop. Dis. 2013, 19, 5. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Progress in the Characterization of Venoms and Standardization of Antivenoms; WHO Offset Publication: Geneva, Switzerland, 1981; pp. 1–44. [Google Scholar]
- Fernandez, J.; Gutierrez, J.M.; Angulo, Y.; Sanz, L.; Juarez, P.; Calvete, J.J.; Lomonte, B. Isolation of an acidic phospholipase A2 from the venom of the snake Bothrops asper of Costa Rica: Biochemical and toxicological characterization. Biochimie 2010, 92, 273–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, V.; Wood, D.L.A.; Newell, F.; Chaumeil, P.-A.; Kaas, Q.; Binford, G.J.; Nicholson, G.M.; Gorse, D.; King, G.F. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2011, 39, D653–D657. [Google Scholar] [CrossRef] [Green Version]
- UniProt. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [Green Version]
- NCBI. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef]
- Pearson, W.R. Finding Protein and Nucleotide Similarities with FASTA. Curr. Protoc. Bioinform. 2016, 53, 3–9. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Ceroni, A.; Passerini, A.; Vullo, A.; Frasconi, P. DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006, 34, W177–W181. [Google Scholar] [CrossRef] [Green Version]
- Ferrè, F.; Clote, P. Disulfide connectivity prediction using secondary structure information and diresidue frequencies. Bioinformatics 2005, 21, 2336–2346. [Google Scholar] [CrossRef] [Green Version]
- Ferrè, F.; Clote, P. DiANNA: A web server for disulfide connectivity prediction. Nucleic Acids Res. 2005, 33, W230–W232. [Google Scholar] [CrossRef] [Green Version]







| A. Phospholipase A2-Like Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c3142 | PhospholipaseA2-1-pverdolaga | Calcium-independent phospholipase A2 | A0A087UHX4 | Stegodyphus mimosarum | 173.33 | 7.09 × 10−50 | 5.79 |
| c4865 | PhospholipaseA2-2-pverdolaga | Cytosolic phospholipase A2 | XP_003214621.2 | Anolis carolinensis | 112.08 | 2.93 × 10−25 | 5.45 |
| c15998 | PhospholipaseA2-3-pverdolaga | Group XV phospholipase A2 | A0A087U096 | S. mimosarum | 560.84 | 0.00 × 10+00 | 28.76 |
| c33599 | PhospholipaseA2-4-pverdolaga | Cytosolic phospholipase A2 | A0A087UL94 | S. mimosarum | 139.04 | 5.33 × 10−39 | 1.51 |
| c45513 | PhospholipaseA2-5-pverdolaga | Calcium-independent phospholipase A2 | A0A087UHX4 | S. mimosarum | 275.02 | 7.49 × 10−88 | 3.09 |
| c10524 | PhospholipaseA2-6-pverdolaga | Calcium-independent phospholipase A2 | A0A0J7L0J5 | Lasius niger | 101.29 | 1.51 × 10−18 | 6.49 |
| c11106 | PhospholipaseA2-7-pverdolaga | Phospholipase A2 | A0A087SVA4 | S. mimosarum | 149.06 | 3.02 × 10−41 | 8.04 |
| c12950 | PhospholipaseA2-8-pverdolaga | Phospholipase A2 | A0A087TLC5 | S. mimosarum | 87.04 | 2.14 × 10−17 | 58.98 |
| c18752 | PhospholipaseA2-9-pverdolaga | Group XIIA secretory phospholipase A2 | XP_011150082.1 | Harpegnathos saltator | 135.58 | 3.87 × 10−35 | 138.53 |
| c21159 | PhospholipaseA2-10-pverdolaga | Calcium-independent phospholipase A2 | E2B1P4 | Camponotus floridanus | 114.39 | 3.47 × 10−24 | 2.68 |
| c42153 | PhospholipaseA2-11-pverdolaga | Cytosolic phospholipase A2 | A0A087UL97 | S. mimosarum | 117.86 | 1.08 × 10−31 | 2.65 |
| c20465 | PhospholipaseA2-12-pverdolaga | Calcium-independent phospholipase A2 | XP_002399324.1 | Ixodes scapularis | 342.81 | 1.19 × 10−105 | 2.66 |
| c29120 | PhospholipaseA2-13-pverdolaga | Group 3 secretory phospholipase A2 | XP_012259279.1 | Athalia rosae | 137.89 | 6.44 × 10−36 | 1.4 |
| c57457 | PhospholipaseA2-14-pverdolaga | Phospholipase A2 | A0A087UYP4 | S. mimosarum | 118.24 | 1.83 × 10−29 | 2.01 |
| c61053 | PhospholipaseA2-15-pverdolaga | Calcium-independent phospholipase A2-γ | A0A087TW26 | S. mimosarum | 479.56 | 1.22 × 10−158 | 10.53 |
| c60448 | PhospholipaseA2-16-pverdolaga | Phospholipase A2-activating protein | B7PWX1 | Ixodes scapularis | 624 | 0.00 × 10+00 | 5.76 |
| B. Phospholipase D-Like Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c46024 | PhospholipaseD-1-pverdolaga | Phospholipase D LiSicTox-betaID1 | Q1W694 | Loxosceles intermedia | 319.70 | 3.17 × 10−100 | 9.24 |
| c14372 | PhospholipaseD-2-pverdolaga | Phospholipase D StSicTox-betaIF1 | C0JB54 | Sicarius terrosus | 214.54 | 4.16 × 10−61 | 16.94 |
| c45658 | PhospholipaseD-3-pverdolaga | Phospholipase D1 | KFM64830.1 | S. mimosarum | 165 | 1 × 10−48 | 1.98 |
| c54699 | PhospholipaseD-4-pverdolaga | Phospholipase D1 | XP_003744259.1 | Metaseiulus occidentalis | 353 | 4.94 × 10−118 | 2.34 |
| C. Phospholipase B-Like Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c17591 | Phospholipase-B-1-pverdolaga | Putative phospholipase B-like 2 | XP_015925352.1 | Parasteatoda tepidariorum | 752 | 0.00 × 10+00 | 31.29 |
| D. KTPSI-Like Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c1808 | Kunitz-1-pverdolaga | Kunitz-type serine protease inhibitor huwentoxin-11g11 | B2ZBB6 | H. schmidti | 75.87 | 9.22 × 10−15 | 5.22 |
| c8989 | Kunitz-2-pverdolaga | Kunitz-type serine protease inhibitor kunitz-1 | W4VSH9 | Trittame loki | 66.24 | 4.76 × 10−10 | 23.43 |
| c12801 | Kunitz-3-pverdolaga | Kunitz-type serine protease inhibitor HWTX-XI-IS4 | P0DJ76 | H. schmidti | 99.37 | 5.62 × 10−23 | 157.37 |
| c15163 | Kunitz-4-pverdolaga | Kunitz-type serine protease inhibitor kunitz-1 | W4VSH9 | Trittame loki | 128.26 | 2.03 × 10−32 | 38.26 |
| c16277 | Kunitz-5-pverdolaga | Kunitz-type serine protease inhibitor 6-like | KFM65460.1 | S. mimosarum | 137.50 | 1 × 10−38 | 13.39 |
| c30159 | Kunitz-6-pverdolaga | Kunitz-type serine protease inhibitor huwentoxin-11g11 | B2ZBB6 | H. schmidti | 92.82 | 4.50 × 10−22 | 76.26 |
| c41726 | Kunitz-7-pverdolaga | Protein with kunitz domain | XP_002435922.1 | Ixodes scapularis | 74.33 | 1.38 × 10−16 | 2.85 |
| c59058 | Kunitz-8-pverdolaga | Kunitz-type serine protease inhibitor huwentoxin-11g11 | B2ZBB6 | H. schmidti | 64.70 | 1.40 × 10−09 | 4.26 |
| c66767 | Kunitz-9-pverdolaga | Kunitz-type protease inhibitor AXPI-I-like | XP_011135446.1 | Harpegnathos saltator | 63.16 | 4.85 × 10−10 | 1.58 |
| c43290 | Kunitz-10-pverdolaga | Kunitz-type protease inhibitor kalicludine-3-like | XP_012273912.1 | Orussus abietinus | 51.99 | 1.71 × 10−06 | 1.66 |
| c52646 | Kunitz-11-pverdolaga | Kunitz-type serine protease inhibitor huwentoxin-11g11 | B2ZBB6 | H. schmidti | 84.73 | 2.64 × 10−17 | 144.34 |
| c6182 | Kunitz-12-pverdolaga | Kunitz-type serine protease inhibitor kunitz-1 | W4VSH9 | Trittame loki | 64.31 | 9.75 × 10−10 | 3.02 |
| c9496 | Kunitz-13-pverdolaga | Kunitz-type serine protease inhibitor huwentoxin-11 | P68425 | H. schmidti | 119.78 | 1.02 × 10−30 | 325.15 |
| E. Hyaluronidase-Like Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c17398 | Hyaluronidase-1-pverdolaga | Hyaluronidase-3 | A0A0F8AST4 | Larimichthys crocea | 79.34 | 4.21 × 10−13 | 14.8 |
| c51925 | Hyaluronidase-2-pverdolaga | Hyaluronidase | J9XYC6 | Brachypelma vagans | 816.99 | 0.00 × 10+00 | 1107.75 |
| F. Lycotoxin-Like Peptides. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c3316 | Lycotoxin-1-pverdolaga | U15-lycotoxin-Ls1d | B6DD42 | Lycosa singoriensis | 50.1 | 6 × 10−06 | 7.36 |
| c28990 | Lycotoxin-2-pverdolaga | U16-lycotoxin-Ls1b | B6DD53 | Lycosa singoriensis | 42 | 8 × 10−7 | 2.35 |
| c13977 | Lycotoxin-3-pverdolaga | U20-lycotoxin-Ls1c-like | A0A087UBG5 | S. mimosarum | 52.37 | 2.96 × 10−06 | 6.38 |
| G. CRISP Proteins. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c9788 | CRISP-1- pverdolaga | GTx-CRISP1 | BAN13537.1 | Grammostola rosea | 518 | 0.00 × 10+00 | 1388.9 |
| C9919 | CRISP-2- pverdolaga | GTx-VA1 | BAN13538.1 | G. rosea | 590 | 0.00 × 10+00 | 18191.29 |
| c18710 | CRISP-3- pverdolaga | GTx-CRISP1 | BAN13537.1 | G. rosea | 322 | 4 × 10−108 | 12.97 |
| H. Hephaestin-Like Protein. | |||||||
| Contig Number | Given Name | Similarity with | Accession number | Organism | Score | E-Value | TPM |
| c5907 | hephaestin-1- pverdolaga | Hephaestin-like protein | XP_021003833.1 | Parasteatoda tepidariorum | 1017 | 0.00 × 10+00 | 4.18 |
| c20814 | hephaestin-2- pverdolaga | Hephaestin | PRD23536.1 | Nephila clavipes | 193 | 1 × 10−59 | 3.16 |
| I. Venom Metalloproteinase. | |||||||
| Contig Number | Given Name | Similarity with | Accession Number | Organism | Score | E-Value | TPM |
| c728 | Metalloproteinase-1- pverdolaga | A disintegrin and metalloproteinase with thrombospondin motifs 1 | KFM63257.1 | S. mimosarum | 758 | 0.00 × 10+00 | 20.99 |
| rp-HPLC | Protein Family | Protein Name | Organism |
|---|---|---|---|
| 6 | Hyaluronidase | Hyaluronidase, partial | Brachypelma vagans |
| 6 | Lycotoxin | U16-lycotoxin-Ls1a | Lycosa singoriensis |
| 21–22 | CRISP | GTx-VA1 | Grammostola rosea |
| CRISP | GTx-VA1 | ||
| CRISP | GTx-VA1 | ||
| CRISP | GTx-VA1 | ||
| 7 | Phospholipases D | Phospholipase D isoform 1 | Loxosceles laeta |
| Phospholipases D | Phospholipase D LlSicTox-alphaIII1i | ||
| 6 | No match | No match | No match |
| Sequences | % Similarity | ID | Match |
|---|---|---|---|
| AGFAGDDAPR | 100 | c6436_g1_i1 | Actin |
| AVFPSIVGRPR | |||
| DSYVGDEAQSKR | |||
| HQGVMVGMGQKDSYVGDEAQSK | |||
| RGILTLK | |||
| EITALAPSTMK | 100 | c62193_g1_i1 | Actin |
| VAPEEHPVLLTEAPLNPK | 100 | c13011_g1_i1 | Actin |
| MTQIMFETFNSPAMYVAIQAVLSLYASGR | 96.55 | ||
| ESRSE | 100 | c15096_g1_i3 | Cytosolic purine 5′-nucleotidase |
| GKPKIQVEYK | 100 | c27174_g1_i1 | Heat shock protein |
| LSKEEIER | 100 | ||
| SENVQDLLLLDVAPLSLGIETAGGVMTALIK | 90.32 | ||
| SENVQDLLLLDVAPLSLGIETAGGVMTSLIK | 90.32 | ||
| GVPQIEVTFDLDANGILQVSAQDKSTGK | 89.29 | ||
| QTQIFTTYSDNQPGVLIQVYEGER | 95.8 | ||
| QTQTFITYSDNQPGVLIQVYEGER | 95.8 | ||
| GVPQIEVTFDIDANGILNVTATDK | 91.67 | ||
| EIAEAYLGYPVTNAVITVPAYFNDSQR | 88.89 | ||
| LLQDFFNGR | 88.89 | ||
| SENVQDLLLLDVAPLSLGLETAGGVMTALIK | 87.1 | ||
| NQVALNPQNTVFDAK | 86.67 | ||
| SENVQDLLLLDVAALSLGLETAGGVMTALIK | 83.87 | ||
| DVLLVDVAPLSLGIETAGGVMTK | 100 | c15743_g1_i1 | Heat shock protein |
| KLFNPEEISAMVLTK | 100 | ||
| LFNPEEISAMVLTK | 100 | ||
| DAGVIAGLNVLR | 91.67 | ||
| TTPSYVAFTDTER | 100 | c10792_g1_i2 | Heat shock protein |
| YRPGTVALREIR | 100 | c2143_g1_i2 | Histone |
| TITLEVEPSDTIENVK | 100 | c16774_g1_i2 | Polyubiquitin-B |
| AGFAGDDAPRAVFPSIVGRPR | 100 | c17180_g4_i1 | Actin |
| DLYANTVLSGGTTMYPGIADRMQK | 100 | ||
| MQKEITALAPSTMK | 100 | ||
| SYELPDGQVITIGNER | 100 | ||
| YSVWIGGSI | 100 | ||
| TTGIVLDSGDGVSHTVPIYEGYALPHAILWLDLAGRDLTDYLMK | 97.73 | ||
| MQKEITALAPSQMK | 92.86 | ||
| MQKEITALAPSWMK | 92.86 | ||
| MQKEITALAPSYMK | 92.86 | ||
| SINPDEAVAYGAAVQAAILMGDK | 95.65 | c16820_g1_i1 | Heat shock protein |
| IINEPTAAALAYGLDR | 93.75 | c9831_g1_i1 | Heat shock protein |
| TITLEVEPSDTAENVK | 93.75 | c16774_g1_i2 | Polyubiquitin-B |
| FELSGIPPAPR | 90.91 | c14336_g1_i1 | Heat shock protein |
| AASSSSTEK | 88.89 | c51913_g1_i1 | Actin |
| MAATKQTAR | 88.89 | c2143_g1_i2 | Histone |
| KSAMATGGVK | 80 | ||
| MAGTKQTAR | 88.89 | c4667_g1_i1 | Histone |
| MANTKQTAR | 88.89 | ||
| AVTKQTAR | 87.5 | ||
| KSAAATGGVK | 80 | ||
| KSACATGGVK | 80 | ||
| KSAEATGGVK | 80 | ||
| KSAGATGGVK | 80 | ||
| KSAHATGGVK | 80 | ||
| KSASATGGVK | 80 | ||
| KSASATGGVK | 80 | ||
| KSAWATGGVK | 80 | ||
| KSAYATGGVK | 80 | ||
| SAIATGGVKKPHR | 84.62 | ||
| CNDMMNVGRLQGFEGK | 87.5 | c15300_g1_i1 | Triple functional domain protein |
| ELEEAER | 85.71 | c34105_g1_i1 | Ubiquitin |
| QAAEAAPEDK | 80 | c5964_g1_i2 | PDZ and LIM domain protein Zasp |
| GSSSGGGYSSGSSSYGSGGR | 80 | c14908_g1_i2 | Protein phosphatase |
| LENEIQTYR | 77.78 | c4688_g1_i1 | Dystonin |
| CKINFCLK | 75 | c14491_g1_i2 | AN1-type zinc finger protein |
| SPATREGK | 75 | c9864_g1_i1 | Cip1-interacting zinc finger protein |
| KVLPLPQR | 75 | c62910_g1_i1 | Developmental protein |
| EDQEQRER | 75 | c29740_g1_i1 | Intersectin-1 |
| KLMEMVNN | 75 | c11572_g1_i1 | Serine/threonine-protein kinase |
| IPCCGKSR | 75 | c5016_g1_i1 | Ubiquitin |
| VQGHSHSK | 75 | c17992_g1_i1 | Uncharacterized protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estrada-Gómez, S.; Vargas-Muñoz, L.J.; Segura Latorre, C.; Saldarriaga-Cordoba, M.M.; Arenas-Gómez, C.M. Analysis of High Molecular Mass Compounds from the Spider Pamphobeteus verdolaga Venom Gland. A Transcriptomic and MS ID Approach. Toxins 2021, 13, 453. https://doi.org/10.3390/toxins13070453
Estrada-Gómez S, Vargas-Muñoz LJ, Segura Latorre C, Saldarriaga-Cordoba MM, Arenas-Gómez CM. Analysis of High Molecular Mass Compounds from the Spider Pamphobeteus verdolaga Venom Gland. A Transcriptomic and MS ID Approach. Toxins. 2021; 13(7):453. https://doi.org/10.3390/toxins13070453
Chicago/Turabian StyleEstrada-Gómez, Sebastian, Leidy Johana Vargas-Muñoz, Cesar Segura Latorre, Monica Maria Saldarriaga-Cordoba, and Claudia Marcela Arenas-Gómez. 2021. "Analysis of High Molecular Mass Compounds from the Spider Pamphobeteus verdolaga Venom Gland. A Transcriptomic and MS ID Approach" Toxins 13, no. 7: 453. https://doi.org/10.3390/toxins13070453
APA StyleEstrada-Gómez, S., Vargas-Muñoz, L. J., Segura Latorre, C., Saldarriaga-Cordoba, M. M., & Arenas-Gómez, C. M. (2021). Analysis of High Molecular Mass Compounds from the Spider Pamphobeteus verdolaga Venom Gland. A Transcriptomic and MS ID Approach. Toxins, 13(7), 453. https://doi.org/10.3390/toxins13070453