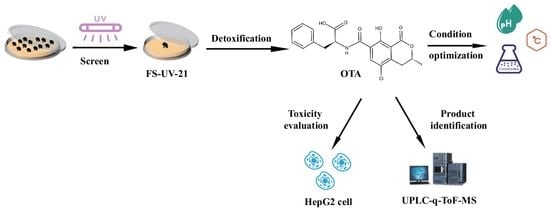
Degradation of Ochratoxin A by a UV-Mutated Aspergillus niger Strain
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening and Identification of the Mutant Strain FS-UV-21
2.2. Degradation of OTA in Liquid Medium by FS10 and FS-UV-21
2.3. Detoxification of Different Concentrations of OTA by the FS-UV-21 Strain and Optimization of Conditions
2.4. Identification of OTA Degradation Products
2.5. OTA Biotransformation Pathway and Metabolic Degradation Products
2.6. Cytotoxicity Evaluation of Degradation Products
2.7. Degradation of OTA by A. niger Biological Fermentation in Wheat Bran
3. Conclusions and Future Perspectives
4. Materials and Methods
4.1. Microorganism and Culture Conditions
4.2. Chemicals
4.3. Preparation of Mutant A. niger Strains
4.4. Identification of the FS-UV-21 Strain
4.5. Detoxification of OTA by FS10 and FS-UV-21
4.6. Extraction and Analysis of OTA
4.7. Effects of Different Factors on the Degradation of OTA by A. niger FS-UV-21
4.8. Extraction and Analysis of OTA Degradation Products
4.9. QTRAP 5500 Combined with LightSight Software Analysis
4.10. Safety Evaluation of OTA Degradation Products
4.11. Bio-Fermentation of A. niger to Degrade OTA in Wheat Bran
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kőszegi, T.; Poór, M. Ochratoxin A: Molecular Interactions, Mechanisms of Toxicity and Prevention at the Molecular Level. Toxins 2016, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mahato, D.K.; Sharma, B.; Borah, R.; Haque, S.; Mahmud, M.C.; Shah, A.K.; Rawal, D.; Bora, H.; Bui, S. Ochratoxins in food and feed: Occurrence and its impact on human health and management strategies. Toxicon 2020, 187, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.D.; Su, P.; Shan, H. Mycotoxin Contamination of Maize in China. Compr. Rev. Food Sci. Food Saf. 2017, 16, 835–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freire, L.; Passamani, F.R.F.; Thomas, A.B.; Nassur, R.D.C.M.R.; Silva, L.M.; Paschoal, F.N.; Pereira, G.E.; Prado, G.; Batista, L. Influence of physical and chemical characteristics of wine grapes on the incidence of Penicillium and Aspergillus fungi in grapes and ochratoxin A in wines. Int. J. Food Microbiol. 2017, 241, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Dai, Q.; Zhao, J.; Qi, X.; Xu, W.; He, X.; Guo, M.; Dweep, H.; Cheng, W.-H.; Luo, Y.; Xia, K.; et al. MicroRNA profiling of rats with ochratoxin A nephrotoxicity. BMC Genom. 2014, 15, 333. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Shen, X.L.; Chen, W.; Liao, X.; Yang, J.; Wang, Y.; Zou, Y.; Fang, C. Advances in research of nephrotoxicity and toxic antagonism of ochratoxin A. Toxin Rev. 2016, 36, 39–44. [Google Scholar] [CrossRef]
- Bui-Klimke, T.R.; Wu, F. Ochratoxin A and Human Health Risk: A Review of the Evidence. Crit. Rev. Food Sci. Nutr. 2013, 55, 1860–1869. [Google Scholar] [CrossRef] [Green Version]
- Czerwiecki, L.; Czajkowska, D.; Witkowska-Gwiazdowska, A. On ochratoxin A and fungal flora in Polish cereals from conventional and ecological farms—Part 1: Occurrence of ochratoxin A and fungi in cereals in 1997. Food Addit. Contam. 2002, 19, 470–477. [Google Scholar] [CrossRef]
- IARC Monographs of the evolution of carcinogenic risks to humans. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; IARC: Lyon, France, 1993. [Google Scholar]
- Wang, L.; Hua, X.; Shi, J.; Jing, N.; Ji, T.; Lv, B.; Liu, L.; Chen, Y. Ochratoxin A: Occurrence and recent advances in detoxification. Toxicon 2022, 210, 11–18. [Google Scholar] [CrossRef]
- Gil-Serna, J.; Vázquez, C.; González-Jaén, M.T.; Patiño, B. Wine Contamination with Ochratoxins: A Review. Beverages 2018, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Beg, M.U.; Al-Mutairi, M.; Beg, K.R.; Al-Mazeedi, H.M.; Ali, L.N.; Saeed, T. Mycotoxins in Poultry Feed in Kuwait. Arch. Environ. Contam. Toxicol. 2006, 50, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Karima, H.-K.; Ridha, G.; Zied, A.; Chekib, M.; Salem, M.; Abderrazek, H. Estimation of Ochratoxin A in human blood of healthy Tunisian population. Exp. Toxicol. Pathol. 2010, 62, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Elling, F.; Nielsen, J.P.; Lillehøj, E.B.; Thomassen, M.; Størmer, F.C. Ochratoxin A-induced porcine nephropathy: Enzyme and ultrastructure changes after short-term exposure. Toxicon 1985, 23, 247–254. [Google Scholar] [CrossRef]
- Shanakhat, H.; Sorrentino, A.; Raiola, A.; Romano, A.; Masi, P.; Cavella, S. Current methods for mycotoxins analysis and innovative strategies for their reduction in cereals: An overview. J. Sci. Food Agric. 2018, 98, 4003–4013. [Google Scholar] [CrossRef]
- Zhu, Y.; Hassan, Y.I.; Lepp, D.; Shao, S.; Zhou, T. Strategies and Methodologies for Developing Microbial Detoxification Systems to Mitigate Mycotoxins. Toxins 2017, 9, 130. [Google Scholar] [CrossRef] [Green Version]
- Varga, J.; Peteri, Z.; Tabori, K.; Teren, J.; Vagvolgyi, C. Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 2005, 99, 321–328. [Google Scholar] [CrossRef]
- Xiong, K.; Zhi, H.-W.; Liu, J.-Y.; Wang, X.-Y.; Zhao, Z.-Y.; Pei, P.-G.; Deng, L.; Xiong, S.-Y. Detoxification of Ochratoxin A by a novel Aspergillus oryzae strain and optimization of its biodegradation. Rev. Argent. Microbiol. 2020, 53, 48–58. [Google Scholar] [CrossRef]
- Bonomo, M.G.; Cafaro, C.; Russo, D.; Calabrone, L.; Milella, L.; Saturnino, C.; Capasso, A.; Salzano, G. Antimicrobial Activity, Antioxidant Properties and Phytochemical Screening of Aesculus hippocastanum Mother Tincture against Food-borne Bacteria. Lett. Drug Des. Discov. 2020, 17, 48–56. [Google Scholar] [CrossRef]
- Sultana, B.; Naseer, R.; Nigam, P. Utilization of agro-wastes to inhibit aflatoxins synthesis by Aspergillus parasiticus: A biotreatment of three cereals for safe long-term storage. Bioresour. Technol. 2015, 197, 443–450. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Inês, A.; Rodrigues, A.; Guimarães, A.; Pereira, V.; Parpot, P.; Mendes-Faia, A.; Venâncio, A. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 2014, 188, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Wang, X.; Xu, S.; Yuan, G.; Shi, X.; Liang, Z. Degradation of ochratoxin A by supernatant and ochratoxinase of Aspergillus niger W-35 isolated from cereals. World Mycotoxin J. 2020, 13, 287–298. [Google Scholar] [CrossRef]
- Campos-Avelar, I.; De La Noue, A.C.; Durand, N.; Fay, B.; Martinez, V.; Fontana, A.; Strub, C.; Schorr-Galindo, S. Minimizing Ochratoxin A Contamination through the Use of Actinobacteria and Their Active Molecules. Toxins 2020, 12, 296. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.R.; Balasubramanian, B.; Park, S.; Jha, R.; Andretta, I.; Bakare, A.G.; Kim, I.H. Ochratoxin A: Carryover from animal feed into livestock and the mitigation strategies. Anim. Nutr. 2020, 7, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Rigó, K.; Téren, J. Degradation of ochratoxin A by Aspergillus species. Int. J. Food Microbiol. 2000, 59, 1–7. [Google Scholar] [CrossRef]
- Bejaoui, H.; Mathieu, F.; Taillandier, P.; Lebrihi, A. Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes: A potential means of ochratoxin A decontamination in grape juices and musts. FEMS Microbiol. Lett. 2006, 255, 203–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Z.L.; Wang, W.; Yang, H.L.; Xia, X.L.; Yu, X.B. Mutation of Acetobacter pasteurianus by UV irradiation under acidic stress for high-acidity vinegar fermentation. Int. J. Food Sci. Tech. 2014, 49, 468–476. [Google Scholar] [CrossRef]
- Zhang, W.B.; Liu, F.N.; Yang, M.; Liang, Q.; Zhang, Y.; Ai, D.Y.; An, Z.G. Enhanced beta-galactosidase production OF Aspergillus oryzae mutated by uv and LiCl. Prep. Biochem. Biotech. 2014, 44, 310–320. [Google Scholar] [CrossRef]
- Zayadan, B.K.; Sadvakasova, A.K.; Userbaeva, A.A.; Bolatkhan, K. Isolation, Mutagenesis, and Optimization of Cultivation Conditions of Microalgal Strains for Biodiesel Production. Russ. J. Plant Physl. 2015, 61, 124–130. [Google Scholar] [CrossRef]
- Kanakdande, A.P.; Khobragade, C.N.; Mane, R.S. Ultraviolet induced random mutagenesis in Bacillus amyloliquefaciens (MF 510169) for improving biodiesel production. Fuel 2021, 304, 121380. [Google Scholar] [CrossRef]
- Yamada, R.; Yamauchi, A.; Kashihara, T.; Ogino, H. Evaluation of lipid production from xylose and glucose/xylose mixed sugar in various oleaginous yeasts and improvement of lipid production by UV mutagenesis. Biochem. Eng. J. 2017, 128, 76–82. [Google Scholar] [CrossRef]
- Xu, D.; Wang, H.; Zhang, Y.; Yang, Z.; Sun, X. Inhibition of non-toxigenic Aspergillus niger FS10 isolated from Chinese fermented soybean on growth and aflatoxin B1 production by Aspergillus flavus. Food Control 2013, 32, 359–365. [Google Scholar] [CrossRef]
- Qiu, T.; Wang, H.; Yang, Y.; Yu, J.; Ji, J.; Sun, J.; Zhang, S.; Sun, X. Exploration of biodegradation mechanism by AFB1-degrading strain Aspergillus niger FS10 and its metabolic feedback. Food Control 2020, 121, 107609. [Google Scholar] [CrossRef]
- Sun, X.; He, X.; Xue, K.S.; Li, Y.; Xu, D.; Qian, H. Biological detoxification of zearalenone by Aspergillus niger strain FS10. Food Chem. Toxicol. 2014, 72, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sun, C.; Zhang, X.; Zhang, H.; Ji, J.; Liu, Y.; Tang, L. Aflatoxin B1 decontamination by UV-mutated live and immobilized Aspergillus niger. Food Control 2016, 61, 235–242. [Google Scholar] [CrossRef]
- Gonzalez-Salgado, A.; Patino, B.; Vazquez, C.; Gonzalez-Jaen, M.T. Discrimination of Aspergillus niger and other Aspergillus species belonging to section Nigri by PCR assays. FEMS Microbiol. Lett. 2005, 245, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Luz, C.; Ferrer, J.; Mañes, J.; Meca, G. Toxicity reduction of ochratoxin A by lactic acid bacteria. Food Chem. Toxicol. 2018, 112, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, M.; Zakowska, Z. The elimination of ochratoxin A by lactic acid bacteria strains. Pol. J. Microbiol. 2005, 54, 279–286. [Google Scholar]
- Chang, X.; Wu, Z.; Wu, S.; Dai, Y.; Sun, C. Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Addit. Contam. Part A 2015, 32, 564–571. [Google Scholar] [CrossRef]
- Shi, L.; Liang, Z.; Li, J.; Hao, J.; Xu, Y.; Huang, K.; Tian, J.; He, X.; Xu, W. Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14. J. Sci. Food Agric. 2013, 94, 1879–1885. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Paterson, R.R.M.; Venâncio, A. Biodegradation of Ochratoxin A for Food and Feed Decontamination. Toxins 2010, 2, 1078–1099. [Google Scholar] [CrossRef] [Green Version]
- Pitout, M.; Nel, W. The inhibitory effect of ochratoxin a on bovine carboxypeptidase a in vitro. Biochem. Pharmacol. 1969, 18, 1837–1843. [Google Scholar] [CrossRef]
- Heussner, A.H.; Bingle, L.E.H. Comparative Ochratoxin Toxicity: A Review of the Available Data. Toxins 2015, 7, 4253–4282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bellis, P.; Tristezza, M.; Haidukowski, M.; Fanelli, F.; Sisto, A.; Mule, G.; Grieco, F. Biodegradation of Ochratoxin A by Bacterial Strains Isolated from Vineyard Soils. Toxins 2015, 7, 5079–5093. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Qian, Y.; Wu, Y.; Chen, Y.; Peng, C.; Luo, M.; Xu, J.; Zhou, Y. Detoxification of ochratoxin A by Lysobacter sp. CW239 and characteristics of a novel degrading gene carboxypeptidase cp4. Environ. Pollut. 2020, 258, 113677. [Google Scholar] [CrossRef]
- el Khoury, A.; Atoui, A. Ochratoxin A: General Overview and Actual Molecular Status. Toxins 2010, 2, 461–493. [Google Scholar] [CrossRef] [Green Version]
- Gayathri, L.; Dhivya, R.; Dhanasekaran, D.; Periasamy, V.S.; Alshatwi, A.; Akbarsha, M.A. Hepatotoxic effect of ochratoxin A and citrinin, alone and in combination, and protective effect of vitamin E: In vitro study in HepG2 cell. Food Chem. Toxicol. 2015, 83, 151–163. [Google Scholar] [CrossRef]
- Prosperini, A.; Juan-García, A.; Font, G.; Ruiz, M. Beauvericin-induced cytotoxicity via ROS production and mitochondrial damage in Caco-2 cells. Toxicol. Lett. 2013, 222, 204–211. [Google Scholar] [CrossRef]
- Haq, M.; Gonzalez, N.; Mintz, K.; Jaja-Chimedza, A.; De Jesus, C.L.; Lydon, C.; Welch, A.Z.; Berry, J.P. Teratogenicity of Ochratoxin A and the Degradation Product, Ochratoxin α, in the Zebrafish (Danio rerio) Embryo Model of Vertebrate Development. Toxins 2016, 8, 40. [Google Scholar] [CrossRef] [Green Version]
- Bittner, A.; Cramer, B.; Harrer, H.; Humpf, H.U. Structure elucidation and in vitro cytotoxicity of ochratoxin alpha amide, a new degradation product of ochratoxin A. Mycotoxin Res. 2015, 31, 83–90. [Google Scholar] [CrossRef]
- Jiang, C.; Shi, J.; Cheng, Y.; Liu, Y. Effect of Aspergillus carbonarius amounts on winemaking and ochratoxin A contamination. Food Control 2014, 40, 85–92. [Google Scholar] [CrossRef]
- Ji, J.; Sun, J.; Pi, F.; Zhang, S.; Sun, C.; Wang, X.; Zhang, Y.; Sun, X. GC-TOF/MS-based metabolomics approach to study the cellular immunotoxicity of deoxynivalenol on murine macrophage ANA-1 cells. Chem. Interact. 2016, 256, 94–101. [Google Scholar] [CrossRef] [PubMed]






| Sample Names | OTA Content (μg/mL) | OTA Degradation Rate (%) | |
|---|---|---|---|
| Fermented 0 d | Fermented 5 d | ||
| Control | 1.142 ± 0.027 | 1.119 ± 0.049 | 2.01 |
| FS-UV-21 | 1.112 ± 0.051 | 0.447 ± 0.064 | 59.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, D.; Ji, J.; Ye, Y.; Yang, Y.; Yu, J.; Wang, M.; Zheng, Y.; Sun, X. Degradation of Ochratoxin A by a UV-Mutated Aspergillus niger Strain. Toxins 2022, 14, 343. https://doi.org/10.3390/toxins14050343
Zou D, Ji J, Ye Y, Yang Y, Yu J, Wang M, Zheng Y, Sun X. Degradation of Ochratoxin A by a UV-Mutated Aspergillus niger Strain. Toxins. 2022; 14(5):343. https://doi.org/10.3390/toxins14050343
Chicago/Turabian StyleZou, Dong, Jian Ji, Yongli Ye, Yang Yang, Jian Yu, Meng Wang, Yi Zheng, and Xiulan Sun. 2022. "Degradation of Ochratoxin A by a UV-Mutated Aspergillus niger Strain" Toxins 14, no. 5: 343. https://doi.org/10.3390/toxins14050343
APA StyleZou, D., Ji, J., Ye, Y., Yang, Y., Yu, J., Wang, M., Zheng, Y., & Sun, X. (2022). Degradation of Ochratoxin A by a UV-Mutated Aspergillus niger Strain. Toxins, 14(5), 343. https://doi.org/10.3390/toxins14050343