Deciphering the Anti-Aflatoxinogenic Properties of Eugenol Using a Large-Scale q-PCR Approach
Abstract
:1. Introduction
2. Results
2.1. Effect of Eugenol on Fungal Growth and Aflatoxin B1 Production
2.2. Effect of Eugenol on Aflatoxin Biosynthetic Pathway
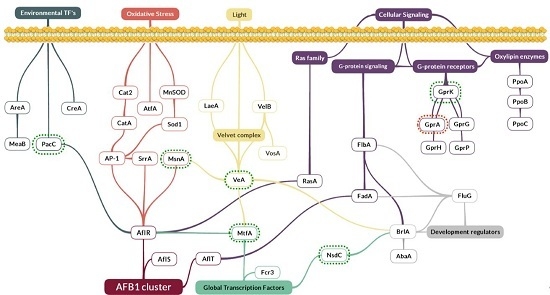
2.3. Effect of Eugenol on Regulatory Factors Linked to AFB1 Production
- The global regulator gene veA, belonging to the velvet complex. It was over-expressed with a 3.8-fold change compared to the control (p-value = 0.002);
- mtfA, a putative C2H2 zinc finger transcription factor. It presented the same up-regulated pattern, increasing its expression by 2.2 times (p-value = 0.0297);
- nsdC, of the global transcription factors, whose expression was increased by 1.7 times (p-value = 0.0100);
- gprK, which was the most affected gene among the five G-protein coupled receptors analyzed here. This gene was over-expressed by 4.5 times (p-value = 0.0009). By contrast, gprA was down-regulated by 0.45 times (p-value = 0.0177);
- The msnA gene was increased by 1.9 times (p-value < 0.0001), whereas no significant changes were observed for other genes implicated in the oxidative stress response such as superoxidase dismutases, catalases or oxylipins;
- Finally, pacC’s expression, a zinc finger transcription factor related to pH, was increased by 2.3 times (p-value = 0.0098).
3. Discussion
3.1. Eugenol Inhibits the Expression of Aflatoxin Cluster Genes in A. flavus
3.2. Eugenol Alters the Expression of Global Regulation Factors
3.2.1. The Pivotal Role of MtfA, VeA and MsnA in Eugenol’s Molecular Mechanism
3.2.2. The Putative Implication of Other Regulatory Factors and Signaling Proteins
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Fungal Strain and Culture Conditions
5.3. Aflatoxin B1 Extraction and Determination by HPLC
5.4. Isolation of Fungal RNA and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
5.5. Design and Validation of q-PCR Primers
5.6. Analysis of the Expression of the Genes Linked to Aflatoxin B1 Biosynthesis
5.7. Statistics
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| A. flavus | Aspergillus flavus |
| AF | Aflatoxins |
| AFB1 | Aflatoxin B1 |
| GPCRs | G-protein coupled receptors |
| HPLC | High Performance Liquid Chromatography |
| MEA | Malt Extract Agar |
| MFS | Major Facilitator Superfamily |
| mM | Millimolar |
| nd | Not detectable |
| ns | No significant changes |
| OE | Over-expresssed |
| ROS | Reactive oxygen species |
| SEM | Standard Error of Mean |
| SM | Secondary metabolism |
| TFs | Transcription factors |
References
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef] [PubMed]
- I.A.R.C. Some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monogr. Eval. Carcinog. Risks Humans. 1993, 56, 245–395. [Google Scholar]
- Bbosa, G.; Kitya, D.; Lubega, A. Review of the biological and health effects of aflatoxins on body organs and body systems. In Aflatoxin—Recent Advances and Future Prospects; INTECH: Hampshire, UK, 2013. [Google Scholar]
- Meissonnier, G.M.; Pinton, P.; Laffitte, J.; Cossalter, A.-M.; Gong, Y.Y.; Wild, C.P.; Bertin, G.; Galtier, P.; Oswald, I.P. Immunotoxicity of aflatoxin B1: Impairment of the cell-mediated response to vaccine antigen and modulation of cytokine expression. Toxicol. Appl. Pharmacol. 2008, 231, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Groopman, J.D.; Kensler, T.W.; Wild, C.P. Protective interventions to prevent aflatoxin-induced carcinogenesis in developing countries. Annu. Rev. Public Health 2008, 29, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Streit, E.; Schatzmayr, G.; Tassis, P.; Tzika, E.; Marin, D.; Taranu, I.; Tabuc, C.; Nicolau, A.; Aprodu, I.; Puel, O.; et al. Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins 2012, 4, 788–809. [Google Scholar] [CrossRef] [PubMed]
- Tabuc, C.; Marin, D.; Guerre, P.; Sesan, T.; Bailly, J.D. Molds and mycotoxin content of cereals in southeastern Romania. J. Food Prot. 2009, 72, 662–665. [Google Scholar] [PubMed]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 2013, 60, 218–237. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.A.; Boston, R.S.; Payne, G.A. Diverse inhibitors of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2008, 78, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: Advantages and limitations. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bluma, R.; Amaiden, M.R.; Daghero, J.; Etcheverry, M. Control of Aspergillus section Flavi growth and aflatoxin accumulation by plant essential oils. J. Appl. Microbiol. 2008, 105, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Kohiyama, C.Y.; Yamamoto Ribeiro, M.M.; Mossini, S.A.G.; Bando, E.; Bomfim, N.D.S.; Nerilo, S.B.; Rocha, G.H.O.; Grespan, R.; Mikcha, J.M.G.; Machinski, M. Antifungal properties and inhibitory effects upon aflatoxin production of Thymus vulgaris L. by Aspergillus flavus Link. Food Chem. 2015, 173, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Shukla, R.; Singh, P.; Dubey, N.K. Chemical composition, antifungal and antiaflatoxigenic activities of Ocimum sanctum L. essential oil and its safety assessment as plant based antimicrobial. Food Chem. Toxicol. 2010, 48, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.D.; Kemmelmeier, C.; Arrotéia, C.C.; da Costa, C.L.; Mallmann, C.A.; Janeiro, V.; Ferreira, F.M.D.; Mossini, S.A.G.; Silva, E.L.; Machinski, M. Inhibitory effect of the essential oil of Curcuma longa L. and curcumin on aflatoxin production by Aspergillus flavus Link. Food Chem. 2013, 136, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-S.; Liu, J.-Y.; Hsui, Y.-R.; Chang, S.-T. Chemical polymorphism and antifungal activity of essential oils from leaves of different provenances of indigenous cinnamon (Cinnamomum osmophloeum). Bioresour. Technol. 2006, 97, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Jahanshiri, Z.; Shams-Ghahfarokhi, M.; Allameh, A.; Razzaghi-Abyaneh, M. Inhibitory effect of eugenol on aflatoxin B1 production in Aspergillus parasiticus by downregulating the expression of major genes in the toxin biosynthetic pathway. World J. Microbiol. Biotechnol. 2015, 31, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Jayashree, T.; Subramanyam, C. Antiaflatoxigenic activity of eugenol is due to inhibition of lipid peroxidation. Lett. Appl. Microbiol. 1999, 28, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Brodhagen, M.; Keller, N.P. Signalling pathways connecting mycotoxin production and sporulation. Mol. Plant. Pathol. 2006, 7, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 2010, 9, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Montibus, M.; Pinson-Gadais, L.; Richard-Forget, F.; Barreau, C.; Ponts, N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 2013, 7828, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Georgianna, D.R.; Payne, G.A. Genetic regulation of aflatoxin biosynthesis: From gene to genome. Fungal Genet. Biol. 2009, 46, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, K.; Carrig, J.; Amare, M.; Keller, N.P. Global survey of canonical Aspergillus flavus G protein-coupled receptors. MBio 2014, 5, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Tsitsigiannis, D.I.; Keller, N.P. Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 2007, 15, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Keller, N.P. Transcriptional regulatory elements in fungal secondary metabolism. J. Microbiol. 2011, 49, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Cary, J.W.; Montalbano, B.G. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Biochim. Biophys. Acta 1999, 1444, 412–417. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Roze, L.; Linz, J. Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism. Toxins (Basel) 2013, 5, 683–702. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Wisecaver, J.H.; Smith, T.D.; Feng, X.; Calvo, A.M.; Rokas, A. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Brooks, W.; Calvo, A. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2003, 2, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Hicks, J.K.; Huang, T.P.; Keller, N.P. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 2003, 165, 1095–1104. [Google Scholar] [PubMed]
- Hicks, J.K.; Yu, J.H.; Keller, N.P.; Adams, T.H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 1997, 16, 4916–4923. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, K.J.; Brodhagen, M.; Keller, N.P. Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins (Basel) 2012, 4, 695–717. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.H.; Hide, W.A.; Yager, L.N.; Lee, B.N. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 1992, 12, 3827–3833. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Mack, B.M.; Wei, Q.; Bland, J.M.; Bhatnagar, D.; Cary, J.W. RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol. Res. 2015, 182, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Baidya, S.; Duran, R.M.; Lohmar, J.M.; Harris-Coward, P.Y.; Cary, J.W.; Hong, S.-Y.; Roze, L.V; Linz, J.E.; Calvo, A.M. VeA is associated with the response to oxidative stress in the aflatoxin producer Aspergillus flavus. Eukaryot. Cell 2014, 13, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, S.; Yoshinari, T.; Furukawa, T.; Jermnak, U.; Takagi, K.; Iimura, K.; Yamamoto, T.; Suzuki, M. Search for aflatoxin and trichothecene production inhibitors and analysis of their modes of action. Biosci. Biotechnol. Biochem. 2016, 80, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Kamatou, G.P.; Vermaak, I.; Viljoen, A.M. Eugenol—From the remote Maluku Islands to the international market place: A review of a remarkable and versatile molecule. Molecules 2012, 17, 6953–6981. [Google Scholar] [CrossRef] [PubMed]
- Chang, P. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genom. 2003, 268, 711–719. [Google Scholar]
- Kong, Q.; Chi, C.; Yu, J.; Shan, S.; Li, Q.; Li, Q.; Guan, B.; Nierman, W.C.; Bennett, J.W. The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2014, 98, 5161–5172. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Campbell, B.C.; Molyneux, R.; Mahoney, N.; Chan, K.L.; Yu, J.; Wilkinson, J.; Cary, J.; Bhatnagar, D.; Cleveland, T.E. Gene targets for fungal and mycotoxin control. Mycotoxin Res. 2006, 22, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, T.; Akiyama, T.; Nakamura, K.; Kondo, T.; Takahashi, Y.; Muraoka, Y.; Nonomura, Y.; Nagasawa, H.; Sakuda, S. Dioctatin A is a strong inhibitor of aflatoxin production by Aspergillus parasiticus. Microbiology 2007, 153, 2774–2780. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C. Predicted roles of the uncharacterized clustered genes in aflatoxin biosynthesis. Toxins (Basel) 2009, 1, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Yu, J.; Yu, J.-H. aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet. Biol. 2004, 41, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Lohmar, J.M.; Satterlee, T.; Cary, J.W.; Calvo, A.M. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins (Basel) 2016, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008, 45, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Bok, J. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004, 70, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Sprote, P.; Brakhage, A.A. The light-dependent regulator velvet A of Aspergillus nidulans acts as a repressor of the penicillin biosynthesis. Arch. Microbiol. 2007, 188, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, S.; Lind, A.L.; Lin, H.-C.; Tang, Y.; Rokas, A.; Calvo, A.M. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Punelli, F.; Camera, E.; Fabbri, C.; Picardo, M.; Fanelli, C.; Fabbri, A.A. Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: A role for the ApyapA gene. Eukaryot. Cell 2008, 7, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Scharfenstein, L.; Luo, M. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins (Basel) 2011, 3, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Roze, L.; Laivenieks, M.; Hong, S.-Y.; Wee, J.; Wong, S.-S.; Vanos, B.; Awad, D.; Ehrlich, K.; Linz, J. Aflatoxin biosynthesis is a novel source of reactive oxygen species—A potential redox signal to initiate resistance to oxidative stress? Toxins (Basel) 2015, 7, 1411–1430. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P.; Nesbitt, C.; Sarr, B.; Phillips, T.D.; Burow, G.B. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology 1997, 87, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W.; Cotty, P.J. Promoter elements in the aflatoxin pathway polyketide synthase gene. Biochim. Biophys. Acta 2002, 1576, 171–175. [Google Scholar] [CrossRef]
- El Mahgubi, A.; Puel, O.; Bailly, S.; Tadrist, S.; Querin, A.; Ouadia, A.; Oswald, I.P.; Bailly, J.D. Distribution and toxigenicity of Aspergillus section Flavi in spices marketed in Morocco. Food Control 2013, 32, 143–148. [Google Scholar] [CrossRef]
- Leite, G.M.; Magan, N.; Medina, Á. Comparison of different bead-beating RNA extraction strategies: An optimized method for filamentous fungi. J. Microbiol. Methods 2012, 88, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Huang, X.; Min, S. Rapid determination of aflatoxins in corn and peanuts. J. Chromatogr. A 2008, 1209, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yang, T. RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol. Biol. Rep. 2002, 20. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caceres, I.; El Khoury, R.; Medina, Á.; Lippi, Y.; Naylies, C.; Atoui, A.; El Khoury, A.; Oswald, I.P.; Bailly, J.-D.; Puel, O. Deciphering the Anti-Aflatoxinogenic Properties of Eugenol Using a Large-Scale q-PCR Approach. Toxins 2016, 8, 123. https://doi.org/10.3390/toxins8050123
Caceres I, El Khoury R, Medina Á, Lippi Y, Naylies C, Atoui A, El Khoury A, Oswald IP, Bailly J-D, Puel O. Deciphering the Anti-Aflatoxinogenic Properties of Eugenol Using a Large-Scale q-PCR Approach. Toxins. 2016; 8(5):123. https://doi.org/10.3390/toxins8050123
Chicago/Turabian StyleCaceres, Isaura, Rhoda El Khoury, Ángel Medina, Yannick Lippi, Claire Naylies, Ali Atoui, André El Khoury, Isabelle P. Oswald, Jean-Denis Bailly, and Olivier Puel. 2016. "Deciphering the Anti-Aflatoxinogenic Properties of Eugenol Using a Large-Scale q-PCR Approach" Toxins 8, no. 5: 123. https://doi.org/10.3390/toxins8050123
APA StyleCaceres, I., El Khoury, R., Medina, Á., Lippi, Y., Naylies, C., Atoui, A., El Khoury, A., Oswald, I. P., Bailly, J. -D., & Puel, O. (2016). Deciphering the Anti-Aflatoxinogenic Properties of Eugenol Using a Large-Scale q-PCR Approach. Toxins, 8(5), 123. https://doi.org/10.3390/toxins8050123