Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators
Abstract
:1. Introduction
2. Results
2.1. LC Production and Purification
2.2. Zymography
2.3. Laccase Identification by MS/MS
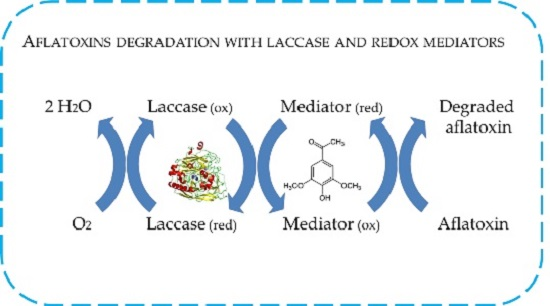
2.4. In Vitro Degradation of AFB1 and AFM1 with Laccase and Redox Mediators
3. Discussion
4. Materials and Methods
4.1. Organism, Culture Conditions, LC Induction, and Production
4.2. Chemicals and Reagents and Standards Preparation
4.3. LC Purification
4.4. LC Spectrophotometric Activity Assay
4.5. Zymography
4.6. LC-MS/MS Analysis
4.7. Bioinformatic Analysis
4.8. In Vitro Degradation of AFB1 and AFM1 with LC and Redox Mediators
4.9. Chemical Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABTS | [2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid)] |
| CAN | Acetonitrile |
| AFB1 | aflatoxin B1 |
| AFM1 | aflatoxin M1 |
| AS | Acetosyringone |
| DEAE | diethylamino ethyl |
| ET | electron transfer |
| EU | enzymatic unit |
| FLD | fluorescent detector |
| HAT | hydrogen atom abstraction |
| HBT | Hydoxybenzotriazole |
| Lac2 | laccase 2 |
| LC | laccase |
| LMS | laccase mediator system |
| LOD | limit of detection |
| LOQ | limit of quantification |
| SA | Syrinagaldehyde |
| TEMPO | 2,2,6,6-tetramethylpiperidine-N-oxyl |
References
- Claus, H. Laccases: Structure, reactions, distribution. Micron 2004, 35, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Osma, J.F.; Toca-Herrera, J.L.; Rodrıguez, S. Uses of Laccases in the Food Industry. Enzym. Res. 2010. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Camarero, S. Laccase engineering by rational and evolutionary design. Cell. Mol. Life Sci. 2015, 72, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Leontievky, A.; Myasoedova, N.; Pozdnyakova, N.; Golovleva, L. “Yellow” laccase of Panus tigrinus oxidizes non-phenolic substrates without redox mediators. FEBS Lett. 2007, 413, 446–448. [Google Scholar] [CrossRef]
- Palmieri, G.; Giardina, P.; Bianco, C.; Scaloni, A.; Capasso, A.; Sannia, G. A novel white laccase from Pleurotus ostreatus. J. Biol. Chem. 1997, 272, 31301–31307. [Google Scholar] [CrossRef] [PubMed]
- Khatun, S.; Islam, S.; Cakilcioglu, U.; Guler, P.; Chatterjee, N.C. Nutritional qualities and antioxidant activity of three edible oyster mushrooms (Pluerotus spp.). NJAS-Wagening. J. Life Sci. 2015, 73, 1–5. [Google Scholar] [CrossRef]
- Munoz, C.; Guillen, F.; Martınez, A.T.; Martınez, M.J. Induction and Characterization of Laccase in the Ligninolytic Fungus Pleurotus eryngii. Curr. Microbiol. 1997, 34, 1–5. [Google Scholar] [PubMed]
- Zucca, P.; Cocco, G.; Sollai, F.; Sanjust, E. Fungal laccases as tools for biodegradation of industrial dyes. Biocatalysis 2015, 1, 82–108. [Google Scholar] [CrossRef] [Green Version]
- Baiocco, P.; Barreca, A.M.; Fabbrini, M.; Galli, C.; Gentili, P. Promoting laccase activity towards non-phenolic substrates: A mechanistic investigation with some laccase-mediator systems. Org. Biomol. Chem. 2003, 1, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Ibarra, D.; Martınez, M.J.; Martınez, A.T. Lignin-Derived Compounds as Efficient Laccase Mediators for Decolorization of Different Types of Recalcitrant Dyes. Appl. Environ. Microbiol. 2005, 71, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Moldes, D.; Díaz, M.; Tzanov, T.; Vidal, T. Comparative study of the efficiency of synthetic and natural mediators in laccase-assisted bleaching of eucalyptus kraft pulp. Bioresour. Technol. 2008, 17, 7959–7965. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Guarino, L.; Piscitelli, A. How to enjoy laccases. Cell. Mol. Life Sci. 2005, 72, 923–940. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.P.; Marth, E.H. Peroxidase activity in mycelia of Aspergillus parasiticus that degrade aflatoxin. Eur. J. Appl. Microbiol. Biotechnol. 1979, 7, 211–217. [Google Scholar] [CrossRef]
- Engelhardt, G. Degradation of Ochratoxin A and B by the White Rot Fungus Pleurotus ostreatus. Mycotoxin Res. 2002, 18, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.F.; Gelderblom, W.C.A.; Botha, A.; van Zyl, W.H. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Banu, I.; Lupu, A.; Aprodu, I. Degradation of Zearalenone by Laccase enzyme. Sci. Study Res. 2013, 14, 79–84. [Google Scholar]
- Aflatoxins. IARC Monographs; International Agency for the Research on Cancer: Lyon, France, 2012; Volume 100F. [Google Scholar]
- Wu, F. Global impacts of aflatoxin in maize: Trade and human health. World Mycotoxin J. 2015, 8, 137–142. [Google Scholar] [CrossRef]
- Dong, J.L.; Zhang, Y.W.; Zhang, R.H.; Huang, W.Z.; Zhang, Y.Z. Influence of culture conditions on laccase production and isozyme patterns in the white-rot fungus Trametes gallica. J. Basic Microbiol. 2005, 45, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Pegler, D.N. The classification of the genus Lentinus Fr. (Basidiomycota). Kavaka 1975, 3, 11–20. [Google Scholar]
- Wu, Q.; Jezkova, A.; Yuan, Z.; Pavlikova, L.; Dohnal, V.; Kuca, K. Biological degradation of aflatoxins. Drug Metab. Rev. 2009, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.H.; Guan, S.; Gao, X.; Ma, Q.G.; Lei, Y.; Bai, X.M.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on Microbial Degradation of Aflatoxins. Crit. Rev. Food Sci. Nutr. 2015. [Google Scholar] [CrossRef] [PubMed]
- Margot, J.; Bennati-Granier, C.; Maillard, J.; Blánquez, P.; Barry, D.A.; Holliger, C. Bacterial versus fungal laccase: Potential for micropollutant degradation. AMB Express 2013, 3, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Scarpari, M.; Bello, C.; Pietricola, C.; Zaccaria, M.; Bertocchi, L.; Angelucci, A.; Ricciardi, M.R.; Scala, V.; Parroni, A.; Fabbri, A.A.; et al. Aflatoxin Control in Maize by Trametes versicolor. Toxins 2014, 6, 3426–3437. [Google Scholar] [CrossRef] [PubMed]
- Zeinvand-Lorestani, H.; Sabzevari, O.; Setayesh, N.; Amini, M.; Nili-Ahmadabadi, A.; Faramarzi, M.A. Comparative study of in vitro prooxidative properties and genotoxicity induced by aflatoxin B1 and its laccase-mediated detoxification products. Chemosphere 2015, 135, 1–6. [Google Scholar] [PubMed]
- Zucca, P.; Rescigno, A.; Olianas, A.; Maccioni, S.; Sollai, F.; Sanjust, E. Induction, purification, and characterization of a laccase isozyme from Pleurotus sajor-caju and the potential in decolorization of textile dyes. J. Mol. Catal. B Enzym. 2011, 68, 216–222. [Google Scholar]
- Díaz, R.; Téllez-Téllez, M.; Sánchez, C.; Bibbins-Martínez, M.D.; Díaz-Godínez, G.; Soriano-Santos, J. Influence of initial pH of the growing medium on the activity, production and genes expression profiles of laccase of Pleurotus ostreatus in submerged fermentation. Electron. J. Biotechnol. 2013, 16. [Google Scholar] [CrossRef]
- Perry, C.R.; Smith, M.; Britnell, C.H.; Wood, D.H.; Thursto, C.F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. Microbiology 1993, 139, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, M.A.; D’Annibale, A.; Galli, C.; Gentili, P.; Sergi, F. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 2008, 6, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Cañas, A.; Camarero, S. Laccases and their natural mediators: Biotechnological tools for sustainable eco-friendly processes. Biotechnol. Adv. 2010, 28, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Bourbonnais, R.; Paice, M.G. Oxidation of non-phenolic substrates: An expanded role for lactase in lignin biodegradation. FEBS Lett. 1990, 267, 99–102. [Google Scholar] [CrossRef]
- Solis-Oba, M.; Almendariz, J.; Viniegra-Gonzalez, G. Biotechnological treatment for colorless denim and textile wastewater. Rev. Int. Contam. Ambient. 2008, 24, 5–11. [Google Scholar]
- Wells, A.; Teria, M.; Eve, T. Green oxidations with laccase-mediator systems. Biochem. Soc. Trans. 2006, 34, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; Kotterman, M.; Field, J.A.; Dobson, A. Oxidation of Anthracene and Benzo [a] pyrene by Laccases from Trametes versicolor. Appl. Environ. Microbiol. 1996, 62, 4563–4567. [Google Scholar] [PubMed]
- Shi, L.; Ma, F.; Han, Y.; Zhang, X.; Yu, H. Removal of sulfonamide antibiotics by oriented immobilized laccase on Fe3O4 nanoparticles with natural mediators. J. Hazard. Mater. 2014, 279, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Dubé, E.; Shareck, F.; Hurtubise, Y.; Beauregard, M.; Daneault, C. Decolourization of recalcitrant dyes with a laccase from Streptomyces coelicolor under alkaline conditions. J. Ind. Microbiol. Biotechnol. 2008, 35, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Ibarra, D.; Martınez, A.T.; Romero, J.; Gutierrez, A.; del Rio, J.C. Paper pulp delignification using laccase and natural mediators. Enzym. Microb. Technol. 2007, 40, 1264–1271. [Google Scholar] [CrossRef] [Green Version]
- Campos, R.; Kandelbauer, A.; Robra, K.H.; Cavaco-Paulo, A.; Guébitz, G.M. Indigo degradation with purified laccases from Trametes hirsuta and Sclerotium rolfsii. J. Biotechnol. 2001, 89, 131–139. [Google Scholar] [CrossRef]
- Rosado, T.; Bernardo, P.; Koci, K.; Coelho, A.V.; Robalo, M.P.; Martins, L.O. Methyl syringate: An efficient phenolic mediator for bacterial and fungal laccases. Bioresour. Technol. 2012, 124, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Elsanhoty, R.M.; Salam, S.A.; Ramadan, M.F.; Badr, F.H. Detoxification of aflatoxin M1 in yoghurt using probiotics and lactic acid bacteria. Food Control 2014, 43, 129–134. [Google Scholar] [CrossRef]
- Vazquez, I.N.; Albores, A.M.; Martınez, E.M.; Miranda, R.; Castro, M. Role of Lactone Ring in Structural, Electronic, and Reactivity Properties of Aflatoxin B1: A Theoretical Study. Arch. Environ. Contam. Toxicol. 2010, 59, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Dunn, J.J.; DeLucca, A.J.; Ciegler, A. Role of lactone ring of aflatoxin B1 in toxicity and mutagenicity. Experientia 1981, 37, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Kersten, P.; Kalyanaraman, B.; Hammel, K.E.; Reinhammar, B.; Kirk, T.K. Comparison of lignin peroxidase, horseradish peroxidase and laccase in the oxidation of methoxybenzenes. Biochem. J. 1990, 268, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Ander, P.; Eriksson, K.E.; Yu, H. Vanillic acid metabolism by Sporotrichum pulverulentum: Evidence for demethoxylation before ring-cleavage. Arch. Clin. Microbiol. 1983, 136, 1–6. [Google Scholar] [CrossRef]
- Commission regulation (EU) 2015/786 of 19 May 2015 defining acceptability criteria for detoxification processes applied to products intended for animal feed as provided for in Directive 2002/32/EC of the European Parliament and of the Council. Off. J. Eur. Union 2015, 58, 10–14.
- ITEM Collection. Available online: http://www.ispa.cnr.it/Collection (accessed on 15 June 2016).
- Association of Official Analytical Chemists (AOAC) Official Method 971.2988 (2000). Available online: http://www.aoacofficialmethod.org/index.php?main_page=product_info&cPath=1&products_id=625 (accessed on 16 August 2016).
- Association of Official Analytical Chemists (AOAC) Official Method 2008.08-2008. Available online: http://www.aoacofficialmethod.org/index.php?main_page=product_info&cPath=1&products_id=2816 (accessed on 16 August 2016).
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitaton of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bleve, G.; Mita, G.; Rampino, P.; Perrotta, C.; Villanova, L.; Grieco, F. Molecular cloning and heterologous expression of a laccase gene from Pleurotus eryngii in free and immobilized Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2008, 79, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Téllez-Téllez, M.; Sánchez, C.; Loera, O.; Díaz-Godínez, G. Differential patterns of constitutive intracellular laccases of the vegetative phase of Pleurotus species. Biotechnol. Lett. 2005, 27, 1391–1394. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Monaci, L.; Pilolli, R.; De Angelis, E.; Godula, M.; Visconti, A. Multi-allergen detection in food by micro high performance liquid chromatography coupled to a dual cell linear ion trap mass spectrometry. J. Chromatogr. A 2014, 1358, 36–144. [Google Scholar] [CrossRef] [PubMed]
- Uniprot Database. Available online: http://www.uniprot.org (accessed on 13 June 2016).




| Purification Step | Total Volume (mL) | Total Activity (EU) | Total Protein (mg) | Specific Activity (U/mg) | Purification Fold |
|---|---|---|---|---|---|
| Crude extract | 4000 | 17,120 | 2800.00 | 6 | 1 |
| Ca-phosphate gel | 500 | 33,400 | 95.00 | 351 | 59 |
| DEAE cellulose | 50 | 23,785 | 11.50 | 2068 | 344 |
| DEAE FF | 10 | 3837 | 1.20 | 3224 | 538 |
| Superdex | 14 | 2053 | 0.19 | 10,920 | 1820 |
| Assigned Protein | Accession Number | Protein Coverage | No. of Identified Peptides | Sequences | Confidence Level | m/z (Da) |
|---|---|---|---|---|---|---|
| Laccase 2 Pleurotus pulmonarius | Q2VT18 | 21.24% | 7 | YSFVLTADQTPDNYWIR | High | 1045.07686 |
| YAGGPTSPLAVINVESTKR | High | 980.65433 | ||||
| SAGSTTYNFDTPAR | High | 744.42450 | ||||
| GDNFQLNVVNQLSDTTMLK | High | 1069.20405 | ||||
| Laccase 4 Pleurotus sajor-caju | Q7Z8S3 | SAGSTTYNFDTPARR | High | 822.55662 | ||
| ANPNLGSTGFAGGINSAILR | High | 644.12372 | ||||
| SVPITGPTPATASIPGVLVQGNK | High | 735.54871 | ||||
| GDNFQLNVVNQLSDTTMLK | High | 718.38048 |
| Sample | AFB1 | AFM1 | |
|---|---|---|---|
| med 1 mM | med 10 mM | med 10 mM | |
| Positive control | 923 ± 33 * | 923 ± 33 * | 53 ± 7 * |
| No med | 710 ± 27 * | 710 ± 27 * | n.t. |
| ABTS | 508 ± 46 * | 175 ± 5 * | 0 ± 0 * |
| AS | 535 ± 9 * | 92 ± 27 * | 0 ± 0 * |
| SA | 803 ± 120 | 258 ± 16 * | 0 ± 0 * |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loi, M.; Fanelli, F.; Zucca, P.; Liuzzi, V.C.; Quintieri, L.; Cimmarusti, M.T.; Monaci, L.; Haidukowski, M.; Logrieco, A.F.; Sanjust, E.; et al. Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins 2016, 8, 245. https://doi.org/10.3390/toxins8090245
Loi M, Fanelli F, Zucca P, Liuzzi VC, Quintieri L, Cimmarusti MT, Monaci L, Haidukowski M, Logrieco AF, Sanjust E, et al. Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins. 2016; 8(9):245. https://doi.org/10.3390/toxins8090245
Chicago/Turabian StyleLoi, Martina, Francesca Fanelli, Paolo Zucca, Vania C. Liuzzi, Laura Quintieri, Maria T. Cimmarusti, Linda Monaci, Miriam Haidukowski, Antonio F. Logrieco, Enrico Sanjust, and et al. 2016. "Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators" Toxins 8, no. 9: 245. https://doi.org/10.3390/toxins8090245
APA StyleLoi, M., Fanelli, F., Zucca, P., Liuzzi, V. C., Quintieri, L., Cimmarusti, M. T., Monaci, L., Haidukowski, M., Logrieco, A. F., Sanjust, E., & Mulè, G. (2016). Aflatoxin B1 and M1 Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins, 8(9), 245. https://doi.org/10.3390/toxins8090245