High-Throughput Gel Microbeads as Incubators for Bacterial Competition Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
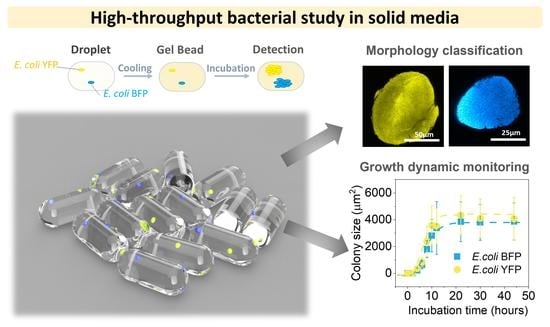
2.2. Encapsulation of Bacteria
2.3. Image Acquisition and Analysis
2.4. Determination of Bacteria Colony Size and Modelling
3. Results and Discussion
3.1. Agarose Microbeads for Bacteria Encapsulation
3.2. Colony Morphology in Agarose Microbeads
3.3. Effect of Inoculum on Colony Size
3.4. Colony Growth Dynamics
3.5. Interactions between E. coli YFP and E. coli BFP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jordán, F.; Lauria, M.; Scotti, M.; Nguyen, T.P.; Praveen, P.; Morine, M.; Priami, C. Diversity of Key Players in the Microbial Ecosystems of the Human Body. Sci. Rep. 2015, 5, 15920. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The Oral Microbiome—An Update for Oral Healthcare Professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, S.; Floury, J.; Gagnaire, V.; Lortal, S.; Thierry, A. Bacterial Colonies in Solid Media and Foods: A Review on Their Growth and Interactions with the Micro-Environment. Front. Microbiol. 2015, 6, 1284. [Google Scholar] [CrossRef] [PubMed]
- Hermans, S.M.; Buckley, H.L.; Case, B.S.; Curran-Cournane, F.; Taylor, M.; Lear, G. Using Soil Bacterial Communities to Predict Physico-Chemical Variables and Soil Quality. Microbiome 2020, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, A.G.; Stephanopoulos, G. Microbial Competition. Science 1981, 213, 972–979. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial Competition: Surviving and Thriving in the Microbial Jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Kim, H.J.; Boedicker, J.; Choi, J.; Ismagilov, R. Defined Spatial Structure Stabilizes a Synthetic Multispecies Bacterial Community. Proc. Natl. Acad. Sci. USA 2008, 105, 18188–18193. [Google Scholar] [CrossRef]
- Monaco, H.; Liu, K.S.; Sereno, T.; Deforet, M.; Taylor, B.P.; Chen, Y.; Reagor, C.C.; Xavier, J.B. Spatial-Temporal Dynamics of a Microbial Cooperative Behavior Resistant to Cheating. Nat. Commun. 2022, 13, 721. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Abdelsamad, A.; Wassermann, T.; Porse, A.; Becker, J.; Sommer, M.O.A.; Høiby, N.; Ciofu, O. The Evolutionary Trajectories of P. Aeruginosa in Biofilm and Planktonic Growth Modes Exposed to Ciprofloxacin: Beyond Selection of Antibiotic Resistance. NPJ Biofilms Microbiomes 2020, 6, 28. [Google Scholar] [CrossRef]
- Siggins, A.; Gunnigle, E.; Abram, F. Exploring Mixed Microbial Community Functioning: Recent Advances in Metaproteomics. FEMS Microbiol. Ecol. 2012, 80, 265–280. [Google Scholar] [CrossRef]
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial Structure, Cooperation and Competition in Biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600. [Google Scholar] [CrossRef]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial Culture through Selective and Non-Selective Conditions: The Evolution of Culture Media in Clinical Microbiology. New Microbes New Infect. 2020, 34, 100622. [Google Scholar] [CrossRef] [PubMed]
- Johnston, T.G.; Yuan, S.F.; Wagner, J.M.; Yi, X.; Saha, A.; Smith, P.; Nelson, A.; Alper, H.S. Compartmentalized Microbes and Co-Cultures in Hydrogels for on-Demand Bioproduction and Preservation. Nat. Commun. 2020, 11, 563. [Google Scholar] [CrossRef]
- Ming, Z.; Han, L.; Bao, M.; Zhu, H.; Qiang, S.; Xue, S.; Liu, W. Living Bacterial Hydrogels for Accelerated Infected Wound Healing. Adv. Sci. 2021, 8, 2102545. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.M.; Barbier, I.; Schaerli, Y. Bacterial Microcolonies in Gel Beads for High-Throughput Screening of Libraries in Synthetic Biology. ACS Synth. Biol. 2017, 6, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Ingham, C.J.; Sprenkels, A.; Bomer, J.; Molenaar, D.; van den Berg, A.; T van Hylckama Vlieg, J.E.; de Vos, W.M. The Micro-Petri Dish, a Million-Well Growth Chip for the Culture and High-Throughput Screening of Microorganisms. Proc. Natl. Acad. Sci. USA 2007, 104, 18217–18222. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, W.; Leng, X.; Zhang, M.; Guan, Z.; Lu, J.; Yang, C.J. Highly Sensitive and Quantitative Detection of Rare Pathogens through Agarose Droplet Microfluidic Emulsion PCR at the Single-Cell Level. Lab Chip 2012, 12, 3907–3913. [Google Scholar] [CrossRef] [PubMed]
- Eun, Y.J.; Utada, A.S.; Copeland, M.F.; Takeuchi, S.; Weibel, D.B. Encapsulating Bacteria in Agarose Microparticles Using Microfluidics for High-Throughput Cell Analysis and Isolation. ACS Chem. Biol. 2011, 6, 260–266. [Google Scholar] [CrossRef]
- Priest, D.G.; Tanaka, N.; Tanaka, Y.; Taniguchi, Y. Micro-Patterned Agarose Gel Devices for Single-Cell High-Throughput Microscopy of E. coli Cells. Sci. Rep. 2017, 7, 17750. [Google Scholar] [CrossRef] [PubMed]
- Krishna Kumar, R.; Meiller-Legrand, T.A.; Alcinesio, A.; Gonzalez, D.; Mavridou, D.A.I.; Meacock, O.J.; Smith, W.P.J.; Zhou, L.; Kim, W.; Pulcu, G.S.; et al. Droplet Printing Reveals the Importance of Micron-Scale Structure for Bacterial Ecology. Nat. Commun. 2021, 12, 857. [Google Scholar] [CrossRef] [PubMed]
- Ceballos-González, C.F.; Bolívar-Monsalve, E.J.; Quevedo-Moreno, D.A.; Lam-Aguilar, L.L.; Borrayo-Montaño, K.I.; Yee-De León, J.F.; Zhang, Y.S.; Alvarez, M.M.; Trujillo-De Santiago, G. High-Throughput and Continuous Chaotic Bioprinting of Spatially Controlled Bacterial Microcosms. ACS Biomater. Sci. Eng. 2021, 7, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Illing, R.; Ruelens, P.; Bachmann, M.; Cuniberti, G.; de Visser, J.A.G.M.; Baraban, L. Coexistence of FluorescentEscherichia Colistrains in Millifluidic Droplet Reactors. Lab Chip 2021, 21, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Baraban, L.; Bertholle, F.; Salverda, M.L.M.; Bremond, N.; Panizza, P.; Baudry, J.; de Visser, J.A.G.M.; Bibette, J. Millifluidic Droplet Analyser for Microbiology. Lab Chip 2011, 11, 4057–4062. [Google Scholar] [CrossRef] [PubMed]
- Baranyi, J.; Roberts, T.A. A Dynamic Approach to Predicting Bacterial Growth in Food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Gopalsamy, K. Exchange of Equilibria in Two Species Lotka-Volterra Competition Models. ANZIAM J. 1982, 24, 160–170. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef]
- Ram, Y.; Dellus-Gur, E.; Bibi, M.; Karkare, K.; Obolski, U.; Feldman, M.W.; Cooper, T.F.; Berman, J.; Hadany, L.; Kerr, B.; et al. Predicting Microbial Growth in a Mixed Culture from Growth Curve Data. Proc. Natl. Acad. Sci. USA 2019, 116, 14698–14707. [Google Scholar] [CrossRef]
- Shao, X.; Mugler, A.; Kim, J.; Jeong, H.J.; Levin, B.R.; Nemenman, I. Growth of Bacteria in 3-d Colonies. PLoS Comput. Biol. 2017, 13, e1005679. [Google Scholar] [CrossRef]
- Buckley, C.T.; Thorpe, S.D.; O’Brien, F.J.; Robinson, A.J.; Kelly, D.J. The Effect of Concentration, Thermal History and Cell Seeding Density on the Initial Mechanical Properties of Agarose Hydrogels. J. Mech. Behav. Biomed. Mater. 2009, 2, 512–521. [Google Scholar] [CrossRef]
- Dell’Arciprete, D.; Blow, M.L.; Brown, A.T.; Farrell, F.D.C.; Lintuvuori, J.S.; McVey, A.F.; Marenduzzo, D.; Poon, W.C.K. A Growing Bacterial Colony in Two Dimensions as an Active Nematic. Nat. Commun. 2018, 9, 4190. [Google Scholar] [CrossRef]
- Skandamis, P.N.; Brocklehurst, T.F.; Panagou, E.Z.; Nychas, G.J.E. Image Analysis as a Mean to Model Growth of Escherichia coli O157:H7 in Gel Cassettes. J. Appl. Microbiol. 2007, 103, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, J.; Xiong, J.Y.; Liu, X.Y. Determination of Agarose Gel Pore Size: Absorbance Measurements Vis a Vis Other Techniques. J. Phys. Conf. Ser. 2006, 28, 83–86. [Google Scholar] [CrossRef]
- Pernodet, N.; Maaloum, M.; Tinland, B. Pore Size of Agarose Gels by Atomic Force Microscopy. Electrophoresis 1997, 18, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Fang, H.H.P. Effective Diffusion Coefficients of Glucose in Artificial Biofilms. Environ. Technol. 2005, 26, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Liang, S.; Zhang, L.; Zhang, X.; Xu, J. Transport of Glucose and Poly(Ethylene Glycol)s in Agarose Gels Studied by the Refractive Index Method. Macromolecules 2005, 38, 5236–5242. [Google Scholar] [CrossRef]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers The Database of Key Numbers in Molecular and Cell Biology. Nucleic Acids Res. 2009, 38, D750–D753. [Google Scholar] [CrossRef]
- Paulton, R.J.L. The Bacterial Growth Curve. J. Biol. Educ. 1991, 25, 92–94. [Google Scholar] [CrossRef]
- Jensen, E.C. Use of Fluorescent Probes: Their Effect on Cell Biology and Limitations. Anat. Rec. 2012, 295, 2031–2036. [Google Scholar] [CrossRef]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a Multidrug Resistance Plasmid by Sublethal Levels of Antibiotics and Heavy Metals. MBio 2014, 5, e01918-14. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen-Le, T.A.; Zhao, X.; Bachmann, M.; Ruelens, P.; Visser, J.A.G.M.d.; Baraban, L. High-Throughput Gel Microbeads as Incubators for Bacterial Competition Study. Micromachines 2023, 14, 645. https://doi.org/10.3390/mi14030645
Nguyen-Le TA, Zhao X, Bachmann M, Ruelens P, Visser JAGMd, Baraban L. High-Throughput Gel Microbeads as Incubators for Bacterial Competition Study. Micromachines. 2023; 14(3):645. https://doi.org/10.3390/mi14030645
Chicago/Turabian StyleNguyen-Le, Trang Anh, Xinne Zhao, Michael Bachmann, Philip Ruelens, J. Arjan G. M. de Visser, and Larysa Baraban. 2023. "High-Throughput Gel Microbeads as Incubators for Bacterial Competition Study" Micromachines 14, no. 3: 645. https://doi.org/10.3390/mi14030645
APA StyleNguyen-Le, T. A., Zhao, X., Bachmann, M., Ruelens, P., Visser, J. A. G. M. d., & Baraban, L. (2023). High-Throughput Gel Microbeads as Incubators for Bacterial Competition Study. Micromachines, 14(3), 645. https://doi.org/10.3390/mi14030645