Spatial Arrangements of Connexin43 in Cancer Related Cells and Re-Arrangements under Treatment Conditions: Investigations on the Nano-Scale by Super-Resolution Localization Light Microscopy
Abstract
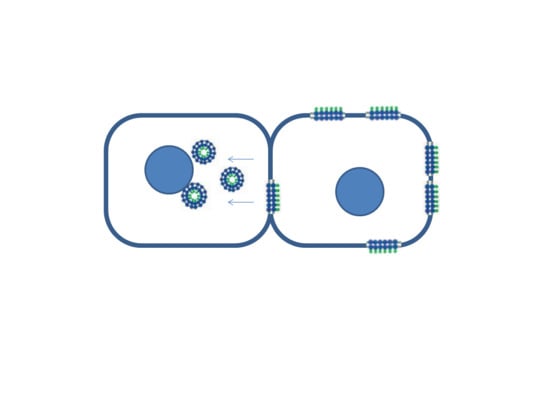
:1. Introduction
2. Results
2.1. SkBr3 Breast Cancer Cells
2.2. Primary Human Internal Mammary Artery Endothelial Cells (HIMAECs)
2.3. Dermal Fibroblasts (BJ cells)
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Treatment and Specimen Preparation
4.2. Labelling and Microscopy
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, J.I.; Wang, L.H. Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. J. Biomed. Sci. 2019, 26, 8. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.V.; Jiang, J.X.; Mesnil, M. Connexins and Pannexins: Important Players in Tumorigenesis, Metastasis and Potential Therapeutics. Int. J. Mol. Sci. 2018, 19, 1645. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Ghosh, S.S. Connexin-43 Enhances the Redesigned Cytosine Deaminase Activity for Suicide Gene Therapy in Human Breast Cancer Cells. Biochem. Insights 2019, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Ghatnekar, G.S.; Yeh, E.S. Connexin 43, breast cancer tumor suppressor: Missed connections? Cancer Lett. 2016, 374, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Pointis, G.; Fiorini, C.; Gilleron, J.; Carette, D.; Segretain, D. Connexins as precocious markers and molecular targets for chemical and pharmacological agents in carcinogenesis. Curr. Med. Chem. 2007, 14, 2288–2303. [Google Scholar] [CrossRef] [PubMed]
- Chasampalioti, M.; Green, A.R.; Ellis, I.O.; Rakha, E.A.; Jackson, A.M.; Spendlove, I.; Ramage, J.M. Connexin 43 is an independent predictor of patient outcome in breast cancer patients. Breast Cancer Res. Treat. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sinyuk, M.; Mulkearns-Hubert, E.E.; Reizes, O.; Lathia, J. Cancer connectors: Connexins, gap junctions, and communication. Front. Oncol. 2018, 8, 646. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Szasz, A.M.; Maros, M.E.; Gyorffy, B.; Kulka, J.; Meggyeshazi, N.; Kiszner, G.; Balla, P.; Samu, A.; Krenacs, T. Correlations of differentially expressed gap junction connexins Cx26, Cx30, Cx32, Cx43 and Cx46 with breast cancer progression and prognosis. PLoS ONE 2014, 9, e112541. [Google Scholar] [CrossRef] [PubMed]
- Bonacquisti, E.E.; Nguyen, J. Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett. 2019, 442, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Heyza, J.R.; Chalfin, E.C.; Ruch, R.J.; Patrick, S.M. Gap Junction Intercellular Communication Positively Regulates Cisplatin Toxicity by Inducing DNA Damage through Bystander Signaling. Cancers (Basel) 2018, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Swietach, P.; Monterisi, S. A Barter Economy in Tumors: Exchanging Metabolites through Gap Junctions. Cancers (Basel) 2019, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Luo, Y.; Mao, N.; Huang, G.; Teng, C.; Wang, H.; Wu, J.; Liao, X.; Yang, J. Cancer-Associated Fibroblasts Accelerate Malignant Progression of Non-Small Cell Lung Cancer via Connexin 43-Formed Unidirectional Gap Junctional Intercellular Communication. Cell Physiol. Biochem. 2018, 51, 315–336. [Google Scholar] [CrossRef] [PubMed]
- Ruch, R.J. Connexin43 suppresses lung cancer stem cells. Cancers (Basel) 2019, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Cha, J.; Kim, S.K.; Park, J.H.; Song, K.H.; Kim, P.; Kim, M.Y. c-MYC Drives Breast Cancer Metastasis to the Brain, but Promotes Synthetic Lethality with TRAIL. Mol. Cancer Res. 2019, 17, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qin, G.; Luo, M.; Chen, J.; Zhang, Q.; Li, L.; Pan, L.; Quin, S. Reciprocal positive regulation between Cx26 and PI3K/Akt pathway confers acquired gefitinib resistance in NSCLC cells via GJIC-independent induction of EMT. Cell Death Diss. 2015, 6, e1829. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Munoz, J.L.; Won, B.W.; Bliss, S.A.; Greco, S.J.; Patel, S.A.; Kandouz, M.; Rameshwar, P. Exogenous CXCL12 activates protein kinase C to phosphorylate connexin 43 for gap junctional intercellular communication among confluent breast cancer cells. Cancer Lett. 2013, 331, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, L.; Fan, L.X.; Huang, K.; Luo, H.M.; Ge, H.; Wang, X.; Wang, Q. Cx32 mediates cisplatin resistance in human ovarian cancer cells by affecting drug efflux transporter expression and activating the EGFR‑Akt pathway. Mol. Med. Rep. 2019, 19, 2287–2296. [Google Scholar] [CrossRef] [PubMed]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2019. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, A.; Rezaei, M.; Niland, S.; Eble, J.A. Collateral Damage Intended-Cancer-Associated Fibroblasts and Vasculature Are Potential Targets in Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2355. [Google Scholar] [CrossRef] [PubMed]
- Elzarrad, M.K.; Haroon, A.; Willeke, K.; Dobrowolski, R.; Gillespie, M.N.; Al-Mehdi, A.B. Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med. 2008, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, G.; Nicolas, V.; Matsusaki, M.; Akashi, M.; Couvreur, P.; Mura, S. Multicellular spheroid based on a triple co-culture: A novel 3D model to mimic pancreatic tumor complexity. Acta Biomater. 2018, 78, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Laschke, M.W.; Menger, M.D. Spheroids as vascularization units: From angiogenesis research to tissue engineering applications. Biotechnol. Adv. 2017, 35, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Chiew, G.G.Y.; Wei, N.; Sultania, S.; Lim, S.; Luo, K.Q. Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: A model system for dual analysis of tumor growth and angiogenesis. Biotechnol. Bioeng. 2017, 114, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.D.; Ding, X.; Zhao, T.; Wu, L.; Perkins, S.; Du, H.; Yan, C. Transthyretin Stimulates Tumor Growth through Regulation of Tumor, Immune, and Endothelial Cells. J. Immunol. 2019, 202, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D. Connexin’s connection in breast cancer growth and progression. Int. J. Cell Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.L.; Williams, C.B.; Zambrano, J.N.; Williams, C.J.; Yeh, E.S. Connexin 43 in the development and progression of breast cancer: What’s the connection? (Review). Int. J. Oncol. 2017, 51, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.S.; Williams, C.J.; Williams, C.B.; Bonilla, I.V.; Klauber-DeNore, N.; Phillips, S.L. Dysregulated connexin 43 in HER2-positive drug resistant breast cancer cells enhances proliferation and migration. Oncotarget 2017, 8, 109358109369. [Google Scholar] [CrossRef] [PubMed]
- Valdebenito, S.; Luo, E.; Baldoni, J.; Okafo, G.; Eugenin, E. The Novel Roles of Connexin Channels and Tunneling Nanotubes in Cancer Pathogenesis. Int. J. Mol. Sci. 2018, 19, 1270. [Google Scholar] [CrossRef] [PubMed]
- Ferrati, S.; Gadok, A.K.; Brunaugh, A.D.; Zhao, C.; Heersema, L.A.; Smyth, H.D.C.; Stachowiak, J.C. Connexin membrane materials as potent inhibitors of breast cancer cell migration. J. R. Soc. Interface. 2017, 133. [Google Scholar] [CrossRef] [PubMed]
- Pilarczyk, G.; Nesnidal, I.; Gunkel, M.; Bach, M.; Bestvater, F.; Hausmann, M. Localisation microscopy of breast epithelial ErbB-2 receptors and gap junctions: Trafficking after gamma-irradiation, Neuregulin-1β and Herceptin application. Int. J. Mol. Sci. 2017, 18, 362. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Mesnil, M.; Naus, C.C.; Lampe, P.D.; Laird, D.W. Gap junctions and cancer: Communicating for 50 years. Nat. Rev. Cancer 2016, 16, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Fallon, R.F.; Goodenough, D.A. Five-hour half-life of mouse liver gap-junction protein. J. Cell Biol. 1981, 90, 521–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, D.W.; Puranam, K.L.; Revel, J.P. Turnover and phosphorylation dynamics of connexin43 gap junction protein in cultured cardiac myocytes. Biochem. J. 1991, 273, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosely, R.; Sorgen, P.L. A history of gap junction structure: hexagonal arrays to atomic resolution. Cell Commun. Adhes. 2013, 20, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Loewenstein, W.R.; Kanno, Y. Intercellular communication and the control of tissue growth: Lack of communication between cancer cells. Nature 1966, 209, 1248–1249. [Google Scholar] [CrossRef] [PubMed]
- Loewenstein, W.R.; Kanno, Y. Intercellular communication and tissue growth. I. Cancerous growth. J. Cell Biol. 1967, 33, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Fentiman, I.S.; Taylor–Papadimitriou, J. Cultured human breast cancer cells lose selectivity in direct intercellular communication. Nature 1977, 269, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Vinken, M.; Decrock, E.; Leybaert, L.; Bultynck, G.; Hompens, B.; Vanhaecke, T.; Rogiers, V. Non-channel functions of connexins in cell growth and cell death. Biophys. Biochim. Acta 2012, 1818, 2002–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhou, Z.; Donahue, H.J. Alterations in Cx43 and OB-cadherin affect breast cancer cell metastatic potential. Clin. Exp. Metastasis 2008, 25, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.M.; Seraj, M.J.; Li, Z.; Zhou, Z.; Winter, C.R.; Welch, D.R.; Donahue, H.J. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001, 61, 1765–1767. [Google Scholar] [PubMed]
- Naus, C.C.; Laird, D.W. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer 2010, 10, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Kandouz, M.; Batist, G. Gap junctions and connexins as therapeutic targets in cancer. Expert Opin. Ther. Targets 2010, 14, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Abt, M.A.; Ghatnekar, G.S.; Yeh, E.S. Targeting connexin 43 with α-connexin carboxylterminal (ACT1) peptide enhances the activity of the targeted inhibitors, tamoxifen and lapatinib, in breast cancer: Clinical implication for ACT1. BMC Cancer 2015, 15, 296. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Leithe, E.; Graham, S.V.; Kameritsch, P.; Mayán, M.D.; Mesnil, M.; Pogoda, K.; Tabernero, A. Connexins in cancer: bridging the gap to the clinic. Oncogene 2019. [Google Scholar] [CrossRef] [PubMed]
- Stoletov, K.; Strnadel, J.; Zardouzian, E.; Momiyama, M.; Park, F.D.; Kelber, J.A.; Pizzo, D.P.; Hoffman, R.; VandenBerg, S.R.; Klemke, R.L. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 2013, 126, 904–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamieson, S.; Going, J.J.; D’Arcy, R.; George, W.D. Expression of gap junction proteins connexin 26 and connexin 43 in normal human breast and in breast tumours. J. Pathol. 1998, 184, 37–43. [Google Scholar] [CrossRef]
- Kańczuga-Koda, L.; Sulkowska, M.; Koda, M.; Reszeć, J.; Famulski, W.; Baltaziak, M.; Sulkowski, S. Expression of connexin 43 in breast cancer in comparison with mammary dysplasia and the normal mammary gland. Folia Morphol (Warsz) 2003, 62, 439–442. [Google Scholar] [PubMed]
- Plante, I.; Stewart, M.K.; Barr, K.; Allan, A.L.; Laird, D.W. Cx43 suppresses mammary tumor metastasis to the lung in a Cx43 mutant mouse model of human disease. Oncogene 2011, 30, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, E.; Shao, Q.; Wang, H.L.; Langlois, S.; Laird, D.W. Connexins act as tumor suppressors in three-dimensional mammary cell organoids by regulating differentiation and angiogenesis. Cancer Res. 2006, 66, 9886–9894. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.; Naczki, C.; Chen, W.; Barlow, K.D.; Case, L.D.; Metheny-Barlow, L.J. Tumor-induced loss of mural Connexin 43 gap junction activity promotes endothelial proliferation. BMC Cancer 2015, 15, 427. [Google Scholar] [CrossRef] [PubMed]
- Zibara, K.; Awada, Z.; Dib, L.; El-Saghir, J.; Al-Ghadban, S.; Ibrik, A.; El-Zein, N.; El-Sabban, M. Anti-angiogenesis therapy and gap junction inhibition reduce MDA-MB-231 breast cancer cell invasion and metastasis in vitro and in vivo. Sci. Rep. 2015, 5, 12598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, C.; Kaufmann, R.; Gunkel, M.; Pres, S.; Weiland, Y.; Müller, P.; Ruckelshausen, T.; Lemmer, P.; Geiger, F.; Degenhard, M.; et al. Superresolution imaging of biological nanostructures by Spectral Precision Distance Microscopy (SPDM). Biotechnol. J. 2011, 6, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Ilić, N.; Pilarczyk, G.; Lee, J.-H.; Logeswaran, A.; Borroni, A.P.; Krufczik, M.; Theda, F.; Waltrich, N.; Bestvater, F.; et al. Challenges for super-resolution localization microscopy and biomolecular fluorescent nano-probing in cancer research. Int. J. Mol. Sci. 2017, 18, 2066. [Google Scholar] [CrossRef] [PubMed]
- Boyd, P.S.; Struve, N.; Bach, M.; Eberle, J.P.; Gote, M.; Schock, F.; Cremer, C.; Kriegs, M.; Hausmann, M. Clustered localization of EGFRvIII in glioblastoma cells as detected by high precision localization microscopy. Nanoscale 2016, 8, 20037–20047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, P.; Lemmermann, N.A.; Kaufmann, R.; Gunkel, M.; Paech, D.; Hildenbrand, G.; Holtappels, R.; Cremer, C.; Hausmann, M. Spatial distribution and structural arrangement of a murine cytomegalovirus glycoprotein detected by SPDM localization microscopy. Histochem. Cell Biol. 2014, 142, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Moser, F.; Hildenbrand, G.; Müller, P.; Al Saroori, A.; Biswas, A.; Bach, M.; Wenz, F.; Cremer, C.; Burger, N.; Veldwijk, M.; et al. Cellular uptake of gold nanoparticles and their behavior as labels for localization microscopy. Biophys. J. 2016, 110, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuß, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-resolution microscopy of radiation-induced histone H2AX phosphorylation in relation to H3K9-trimethylation in HeLa cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef] [PubMed]
- Eryilmaz, M.; Schmitt, E.; Krufczik, M.; Theda, F.; Lee, J.-H.; Cremer, C.; Bestvater, F.; Schaufler, W.; Hausmann, M.; Hildenbrand, G. Localization microscopy analyses of MRE11 clusters in 3D-conserved cell nuclei of different cell lines. Cancers 2018, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Bach, M.; Savini, C.; Krufczik, M.; Cremer, C.; Rösl, F.; Hausmann, M. Super-resolution localization microscopy of γ-H2AX and heterochromatin after folate deficiency. Int. J. Mol. Sci. 2017, 18, 1726. [Google Scholar] [CrossRef] [PubMed]
- Lemmer, P.; Gunkel, M.; Baddeley, D.; Kaufmann, R.; Urich, A.; Weiland, Y.; Reymann, J.; Müller, P.; Hausmann, M.; Cremer, C. SPDM—Light microscopy with single molecule resolution at the nanoscale. Appl. Phys. B 2018, 93, 1–12. [Google Scholar] [CrossRef]
- Lemmer, P.; Gunkel, M.; Weiland, Y.; Müller, P.; Baddeley, D.; Kaufmann, R.; Urich, A.; Eipel, H.; Amberger, R.; Hausmann, M.; et al. Using conventional fluorescent markers for far-field fluorescence localization nanoscopy allows resolution in the 10 nm range. J. Microsc. 2009, 235, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Ripley, B.D. Modeling Spatial Patterns. J Royal Statist Soc B (Methodological) 1977, 39, 172–212. [Google Scholar] [CrossRef]
- Giepmans, B.N.G. Gap junctions and connexin-interacting proteins. Cardiovasc. Res. 2004, 62, 233–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severs, N.J.; Dupont, E.; Coppen, S.R.; Halliday, D.; Inett, E.; Baylis, D.; Rothery, S. Remodeling of gap junctions and connexin expression in heart disease. Biochim. Biophys. Acta 2004, 1662, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Kostin, S.; Hein, S.; Bauer, E.P.; Schaper, J. Spatiotemporal development and distribution of intercellular junctions in adult rat cardiac myocytes in culture. Circ. Res. 1999, 85, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Zhuo, X.Z.; Ni, Y.J.; Gong, M.; Wang, T.Z.; Lu, Q.; Ma, A.Q. Improvement of cardiac function and reversal of gap junction remodeling by neuregulin-1β in volume-overloaded rats with heart failure. J. Geriatr. Cardiol. 2012, 9, 172–179. [Google Scholar] [PubMed]
- Gu, Y.; Huang, F.; Wang, Y.; Chen, C.; Wu, S.; Zhou, S.; Hei, Z.; Yuan, D. Connexin32 plays a crucial role in ROS-mediated endoplasmic reticulum stress apoptosis signaling pathway in ischemia reperfusion-induced acute kidney injury. J. Transl. Med. 2018, 16, 117. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.D.; Camelitti, P. Role of Non-Myocyte Gap Junctions and Connexin Hemichannels in Cardiovascular Health and Disease: Novel Therapeutic Targets? Int. J. Mol. Sci. 2018, 19, 866. [Google Scholar] [CrossRef] [PubMed]
- Krufczik, M.; Sievers, A.; Hausmann, A.; Lee, J.-H.; Hildenbrand, G.; Schaufler, W.; Hausmann, M. Combining low temperature fluorescence DNA-hybridization, immunostaining, and super-resolution localization microscopy for nano-structure analysis of ALU elements and their influence on chromatin structure. Int. J. Mol. Sci. 2017, 18, 1005. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmüller, M.; Schwarz-Finsterle, J.; Fey, E.; Lux, J.; Bach, M.; Cremer, C.; Hinderhofer, K.; Hausmann, M.; Hildenbrand, G. In situ optical sequencing and nano-structure analysis of a trinucleotide expansion region by localization microscopy after specific COMBO-FISH labelling. Nanoscale 2015, 7, 17938–17946. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.; Krufczik, M.; Heermann, D.W.; Hausmann, M. Using persistent homology as a new approach for super-resolution localization microscopy data analysis and classification of H2AX foci/clusters. Int. J. Mol. Sci. 2018, 19, 2263. [Google Scholar] [CrossRef] [PubMed]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilarczyk, G.; Papenfuß, F.; Bestvater, F.; Hausmann, M. Spatial Arrangements of Connexin43 in Cancer Related Cells and Re-Arrangements under Treatment Conditions: Investigations on the Nano-Scale by Super-Resolution Localization Light Microscopy. Cancers 2019, 11, 301. https://doi.org/10.3390/cancers11030301
Pilarczyk G, Papenfuß F, Bestvater F, Hausmann M. Spatial Arrangements of Connexin43 in Cancer Related Cells and Re-Arrangements under Treatment Conditions: Investigations on the Nano-Scale by Super-Resolution Localization Light Microscopy. Cancers. 2019; 11(3):301. https://doi.org/10.3390/cancers11030301
Chicago/Turabian StylePilarczyk, Götz, Franziska Papenfuß, Felix Bestvater, and Michael Hausmann. 2019. "Spatial Arrangements of Connexin43 in Cancer Related Cells and Re-Arrangements under Treatment Conditions: Investigations on the Nano-Scale by Super-Resolution Localization Light Microscopy" Cancers 11, no. 3: 301. https://doi.org/10.3390/cancers11030301
APA StylePilarczyk, G., Papenfuß, F., Bestvater, F., & Hausmann, M. (2019). Spatial Arrangements of Connexin43 in Cancer Related Cells and Re-Arrangements under Treatment Conditions: Investigations on the Nano-Scale by Super-Resolution Localization Light Microscopy. Cancers, 11(3), 301. https://doi.org/10.3390/cancers11030301