Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
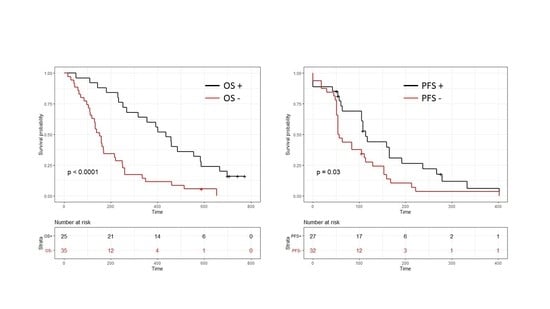
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients and Sample Collection
4.2. Gene Expression Analysis via qPCR
4.3. Statistical Analysis: Selection of Candidate Genes
4.4. Statistical Analysis: Gene Expression-Based (GE) Score
4.5. Statistical Analysis: Univariate and Multivariate Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Neuzillet, C.; Gaujoux, S.; Williet, N.; Bachet, J.B.; Bauguion, L.; Colson Durand, L.; Conroy, T.; Dahan, L.; Gilabert, M.; Huguet, F.; et al. Pancreatic cancer: French clinical practice guidelines for diagnosis, treatment and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO, ACHBT, AFC). Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2018, 50, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Partensky, C.; Bray, F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016, 55, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.S.; Cheung, W.Y. A Contemporary Review of the Treatment Landscape and the Role of Predictive and Prognostic Biomarkers in Pancreatic Adenocarcinoma. Can. J. Gastroenterol. Hepatol. 2018, 2018, 1863535. [Google Scholar] [CrossRef] [Green Version]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, A.M.; Hidalgo, M.; Alvarez, R.; Arrazubi, V.; Martinez-Galan, J.; Salgado, M.; Macarulla, T.; Carrato, A. From First Line to Sequential Treatment in the Management of Metastatic Pancreatic Cancer. J Cancer 2018, 9, 1978–1988. [Google Scholar] [CrossRef] [PubMed]
- Raffenne, J.; Nicolle, R.; Puleo, F.; Le Corre, D.; Boyez, C.; Marechal, R.; Emile, J.F.; Demetter, P.; Bardier, A.; Laurent-Puig, P.; et al. hENT1 Testing in Pancreatic Ductal Adenocarcinoma: Are We Ready? A Multimodal Evaluation of hENT1 Status. Cancers 2019, 11, 1808. [Google Scholar] [CrossRef] [Green Version]
- Puleo, F.; Nicolle, R.; Blum, Y.; Cros, J.; Marisa, L.; Demetter, P.; Quertinmont, E.; Svrcek, M.; Elarouci, N.; Iovanna, J.; et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018, 155, 1999–2013.e3. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.M.; Yoon, K.A.; Hong, E.K.; Park, W.S.; Han, S.S.; Park, S.J.; Joo, J.; Park, E.Y.; Lee, J.H.; Kim, Y.H.; et al. DCK expression, a potential predictive biomarker in the adjuvant gemcitabine chemotherapy for biliary tract cancer after surgical resection: Results from a phase II study. Oncotarget 2017, 8, 81394–81404. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, L.; Iacobazzi, R.M.; Di Fonte, R.; Serrati, S.; Intini, A.; Solimando, A.G.; Brunetti, O.; Calabrese, A.; Leonetti, F.; Azzariti, A.; et al. CAFs and TGF-beta Signaling Activation by Mast Cells Contribute to Resistance to Gemcitabine/Nabpaclitaxel in Pancreatic Cancer. Cancers 2019, 11, 330. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Pant, S.; Ryan, D.P.; Laheru, D.; Bahary, N.; Dragovich, T.; Hosein, P.J.; Rolfe, L.; Saif, M.W.; LaValle, J.; et al. A phase II, open-label, multicenter study to evaluate the antitumor efficacy of CO-1.01 as second-line therapy for gemcitabine-refractory patients with stage IV pancreatic adenocarcinoma and negative tumor hENT1 expression. Pancreatology 2014, 14, 398–402. [Google Scholar] [CrossRef] [Green Version]
- Bachet, J.B.; Blons, H.F.; Hammel, P.; El Hariry, I.; Portales, F.; Mineur, L.; Metges, J.P.; Mulot, C.; Bourreau, C.; Cain, J.; et al. Circulating tumor DNA is prognostic and potentially predictive of eryaspase efficacy in second-line in patients with advanced pancreatic adenocarcinoma. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Deplanque, G.; Demarchi, M.; Hebbar, M.; Flynn, P.; Melichar, B.; Atkins, J.; Nowara, E.; Moye, L.; Piquemal, D.; Ritter, D.; et al. A randomized, placebo-controlled phase III trial of masitinib plus gemcitabine in the treatment of advanced pancreatic cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1194–1200. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Honda, M.; Matsui, S.; Komori, O.; Murayama, T.; Fujiwara, T.; Mizuno, M.; Imai, Y.; Yoshimura, K.; Nasti, A.; et al. Development of novel diagnostic system for pancreatic cancer, including early stages, measuring mRNA of whole blood cells. Cancer Sci. 2019, 110, 1364–1388. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Peng, Y.; Zhu, Y.; Xu, D.; Zhu, F.; Xu, W.; Chen, Q.; Zhu, X.; Liu, T.; Hou, C.; et al. Glycolysis promotes the progression of pancreatic cancer and reduces cancer cell sensitivity to gemcitabine. Biomed. Pharm. 2020, 121, 109521. [Google Scholar] [CrossRef]
- Jia, Y.; Xie, J. Promising molecular mechanisms responsible for gemcitabine resistance in cancer. Genes Dis. 2015, 2, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Xue, X.; Wang, F.; An, Y.; Tang, D.; Xu, Y.; Wang, H.; Yuan, Z.; Gao, W.; Wei, J.; et al. Expression and promoter methylation analysis of ATP-binding cassette genes in pancreatic cancer. Oncol. Rep. 2012, 27, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Hastie, T. Regularization and variable selection via theelastic net. J. R Stat. Soc. B 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Hoerl, A.; Kennard, R. Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Mellert, H.; Foreman, T.; Jackson, L.; Maar, D.; Thurston, S.; Koch, K.; Weaver, A.; Cooper, S.; Dupuis, N.; Sathyanarayana, U.G.; et al. Development and Clinical Utility of a Blood-Based Test Service for the Rapid Identification of Actionable Mutations in Non-Small Cell Lung Carcinoma. J. Mol. Diagn. 2017, 19, 404–416. [Google Scholar] [CrossRef] [Green Version]

| Analyze | Univariate | Multivariate | ||
|---|---|---|---|---|
| Result | HR (95% CI for HR) | p-Value | HR (95% CI for HR) | p-Value |
| GE score OS + prediction | 0.31 (0.17–0.56) | 0.000095 | 0.39 (0.21–0.7) | 0.002 |
| CA 19–9 (U/mL) | 1 (1–1) | 0.28 | ||
| Albumin (g/L) | 0.97 (0.94–1) | 0.16 | ||
| QLQ-C30 | 1 (1–1) | 0.0027 | 1.02 (1.0–1.04) | 0.015 |
| Body mass index | 0.98 (0.92–1) | 0.5 | ||
| ECOG PS | 1.8 (0.96–3.2) | 0.067 | ||
| Monocyte count (per µL) | 2 (0.9–4.6) | 0.09 | ||
| Tumor localization | ||||
| Head | 0.86 (0.51–1.5) | 0.59 | ||
| Body | 1.1 (0.62–1.9) | 0.81 | ||
| Tail | 1.5 (0.85–2.7) | 0.16 | ||
| Clinical stage | 0.34 (0.17–0.68) | 0.0024 | 0.41 (0.2–0.83) | 0.014 |
| Analyze | Univariate | Multivariate | ||
|---|---|---|---|---|
| Result | HR (95% CI for HR) | p-Value | HR (95% CI for HR) | p-Value |
| GE score PFS + prediction | 0.55 (0.32–0.95) | 0.032 | 0.5 (0.28–0.9) | 0.025 |
| CA 19–9 (U/mL) | 1 (1–1) | 0.47 | ||
| Albumin (g/L) | 1 (0.96–1) | 0.81 | ||
| QLQ-C30 | 1 (1–1) | 0.038 | 1.02 (1.0–1.04) | 0.026 |
| Body mass index | 0.99 (0.93–1.1) | 0.76 | ||
| ECOG PS | 2 (1–3.8) | 0.045 | 1.6 (0.8–3.1) | 0.17 |
| Monocyte count (per µL) | 0.73 (0.29–1.8) | 0.49 | ||
| Tumor localization | ||||
| head | 1.2 (0.69–2.1) | 0.52 | ||
| body | 1.1 (0.64–1.9) | 0.7 | ||
| tail | 1.5 (0.84–2.8) | 0.16 | ||
| Clinical classification | 0.7 (0.35–1.4) | 0.32 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piquemal, D.; Noguier, F.; Pierrat, F.; Bruno, R.; Cros, J. Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer. Cancers 2020, 12, 3204. https://doi.org/10.3390/cancers12113204
Piquemal D, Noguier F, Pierrat F, Bruno R, Cros J. Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer. Cancers. 2020; 12(11):3204. https://doi.org/10.3390/cancers12113204
Chicago/Turabian StylePiquemal, David, Florian Noguier, Fabien Pierrat, Roman Bruno, and Jerome Cros. 2020. "Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer" Cancers 12, no. 11: 3204. https://doi.org/10.3390/cancers12113204
APA StylePiquemal, D., Noguier, F., Pierrat, F., Bruno, R., & Cros, J. (2020). Predictive Values of Blood-Based RNA Signatures for the Gemcitabine Response in Advanced Pancreatic Cancer. Cancers, 12(11), 3204. https://doi.org/10.3390/cancers12113204