Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis
Abstract
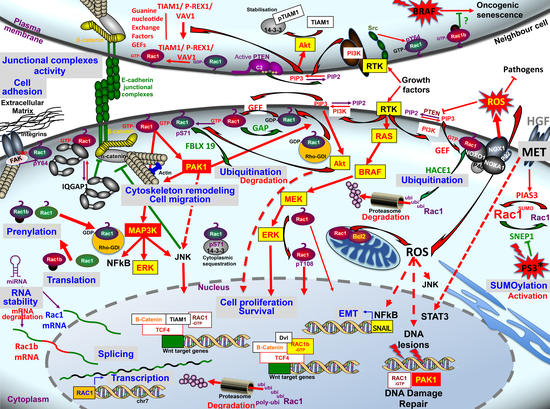
:1. Introduction
2. Rac1 in Intestinal Physiology
2.1. Role of Rac1 in Intestinal Homeostasis
2.2. Intestinal Barrier Integrity/Mucosal Repair/Colitis
3. Rac1 in Colorectal Carcinogenesis
3.1. Rac1 Overexpression and Activation in CRC
3.2. Rac1 Post Translational Modifications (PTMs) in CRC
3.2.1. Rac1 SUMOylation
3.2.2. Rac1 Ubiquitination
3.2.3. Rac1 Phosphorylation
3.3. RAC1b Splice Variant
3.4. Rac1 Regulators & Effectors
3.4.1. Guanine nucleotide Exchange Factors (GEFs)
3.4.2. GTPase-Activating Proteins (GAP)/Rho GDP Dissociation Inhibitor (RhoGDI)
3.4.3. Rac1 Effectors and Signaling Pathways
4. Conclusions, Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Ait Ouakrim, D.; Pizot, C.; Boniol, M.; Malvezzi, M.; Boniol, M.; Negri, E.; Bota, M.; Jenkins, M.A.; Bleiberg, H.; Autier, P. Trends in colorectal cancer mortality in Europe: Retrospective analysis of the WHO mortality database. BMJ 2015, 351, h4970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Kotelevets, L.; Chastre, E.; Desmaële, D.; Couvreur, P. Nanotechnologies for the treatment of colon cancer: From old drugs to new hope. Int. J. Pharm. 2016, 514, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Svensmark, J.H.; Brakebusch, C. Rho GTPases in cancer: Friend or foe? Oncogene 2019, 38, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Aske, J.C.; Dey, N. RAC1 Takes the Lead in Solid Tumors. Cells 2019, 8, 382. [Google Scholar] [CrossRef] [Green Version]
- Marei, H.; Carpy, A.; Woroniuk, A.; Vennin, C.; White, G.; Timpson, P.; Macek, B.; Malliri, A. Differential Rac1 signalling by guanine nucleotide exchange factors implicates FLII in regulating Rac1-driven cell migration. Nat. Commun. 2016, 7, 10664. [Google Scholar] [CrossRef] [Green Version]
- Marei, H.; Carpy, A.; Macek, B.; Malliri, A. Proteomic analysis of Rac1 signaling regulation by guanine nucleotide exchange factors. Cell Cycle 2016, 15, 1961–1974. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Gao, Y.; Moon, S.Y.; Zhang, Y.; Zheng, Y. Oligomerization of Rac1 gtpase mediated by the carboxyl-terminal polybasic domain. J. Biol. Chem. 2001, 276, 8958–8967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casado-Medrano, V.; Baker, M.J.; Lopez-Haber, C.; Cooke, M.; Wang, S.; Caloca, M.J.; Kazanietz, M.G. The role of Rac in tumor susceptibility and disease progression: From biochemistry to the clinic. Biochem. Soc. Trans. 2018, 46, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Brazåo, R.; Boavida, M.G.; Gespach, C.; Chastre, E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999, 18, 6835–6839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotelevets, L.; Walker, F.; Mamadou, G.; Lehy, T.; Jordan, P.; Chastre, E. The Rac1 splice form Rac1b favors mouse colonic mucosa regeneration and contributes to intestinal cancer progression. Oncogene 2018, 37, 6054–6068. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Hass, R.; Lehnert, H.; Ungefroren, H. RAC1B: A Rho GTPase with Versatile Functions in Malignant Transformation and Tumor Progression. Cells 2019, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Kwon, T.; Kwon, D.Y.; Chun, J.; Kim, J.H.; Kang, S.S. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J. Biol. Chem. 2000, 275, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Kotelevets, L.; Scott, M.G.H.; Chastre, E. Targeting PTEN in Colorectal Cancers. Adv. Exp. Med. Biol. 2018, 1110, 55–73. [Google Scholar]
- Wang, F.-S.; Wu, W.-H.; Hsiu, W.-S.; Liu, Y.-J.; Chuang, K.-W. Genome-Scale Metabolic Modeling with Protein Expressions of Normal and Cancerous Colorectal Tissues for Oncogene Inference. Metabolites 2020, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation of Rac1 T108 by extracellular signal-regulated kinase in response to epidermal growth factor: A novel mechanism to regulate Rac1 function. Mol. Cell. Biol. 2013, 33, 4538–4551. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- García-Aranda, M.; Redondo, M. Targeting Receptor Kinases in Colorectal Cancer. Cancers 2019, 11, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, F.; Lemmon, C.; Lietha, D.; Eck, M.; Romer, L. Tyrosine Phosphorylation of Rac1: A Role in Regulation of Cell Spreading. PLoS ONE 2011, 6, e28587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frame, M.C. Newest findings on the oldest oncogene how activated src does it. J. Cell Sci. 2004, 117, 989–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aljagthmi, A.A.; Hill, N.T.; Cooke, M.; Kazanietz, M.G.; Abba, M.C.; Long, W.; Kadakia, M.P. ΔNp63α suppresses cells invasion by downregulating PKCγ/Rac1 signaling through miR-320a. Cell Death Dis. 2019, 10, 680. [Google Scholar] [CrossRef]
- Parsons, M.; Adams, J.C. Rac regulates the interaction of fascin with protein kinase C in cell migration. J. Cell Sci. 2008, 121, 2805–2813. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.M.A.; Patel, R.; Acevedo-Duncan, M. Protein Kinase C-ζ stimulates colorectal cancer cell carcinogenesis via PKC-ζ/Rac1/Pak1/β-Catenin signaling cascade. Cell Res. 2018, 1865, 650–664. [Google Scholar]
- Kang, H.T.; Park, J.T.; Choi, K.; Choi, H.J.C.; Jung, C.W.; Kim, G.R.; Lee, Y.-S.; Park, S.C. Chemical screening identifies ROCK as a target for recovering mitochondrial function in Hutchinson-Gilford progeria syndrome. Aging Cell 2017, 16, 541–550. [Google Scholar] [CrossRef]
- Cai, S.-D.; Chen, J.-S.; Xi, Z.-W.; Zhang, L.-J.; Niu, M.-L.; Gao, Z.-Y. MicroRNA-144 inhibits migration and proliferation in rectal cancer by downregulating ROCK-1. Mol. Med. Rep. 2015, 12, 7396–7402. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Kenney, S.R.; Muller, C.Y.; Adams, S.; Rutledge, T.; Romero, E.; Murray-Krezan, C.; Prekeris, R.; Sklar, L.A.; Hudson, L.G.; et al. R-Ketorolac Targets Cdc42 and Rac1 and Alters Ovarian Cancer Cell Behaviors Critical for Invasion and Metastasis. Mol. Cancer Ther. 2015, 14, 2215–2227. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Lluva, S.; Tatham, M.H.; Jones, R.C.; Jaffray, E.G.; Edmondson, R.D.; Hay, R.T.; Malliri, A. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 2010, 12, 1078–1085. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Fu, Y.; Guo, F.; Zhang, X.; Xiao, S.; Zhu, W.; Huang, Z.; Zhang, J.; Chen, J. PIAS3-mediated feedback loops promote chronic colitis-associated malignant transformation. Theranostics 2018, 8, 3022–3037. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Zhang, C.; Zhao, Y.; Liu, J.; Lin, A.W.; Tan, V.M.; Drake, J.M.; Liu, L.; Boateng, M.N.; Li, J.; et al. Gain-of-function mutant p53 activates small GTPase Rac1 through SUMOylation to promote tumor progression. Genes Dev. 2017, 31, 1641–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo-Lluva, S.; Tan, C.-T.; Daugaard, M.; Sorensen, P.H.B.; Malliri, A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2013, 32, 1735–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugaard, M.; Nitsch, R.; Razaghi, B.; McDonald, L.; Jarrar, A.; Torrino, S.; Castillo-Lluva, S.; Rotblat, B.; Li, L.; Malliri, A.; et al. Hace1 controls ROS generation of vertebrate Rac1-dependent NADPH oxidase complexes. Nat. Commun. 2013, 4, 2180. [Google Scholar] [CrossRef] [Green Version]
- Acosta, M.I.; Urbach, S.; Doye, A.; Ng, Y.-W.; Boudeau, J.X.R.X.M.; Mettouchi, A.; Debant, A.; Manser, E.; Visvikis, O.; Lemichez, E. Group-I PAKs-mediated phosphorylation of HACE1 at serine 385 regulates its oligomerization state and Rac1 ubiquitination. Sci. Rep. 2018, 8, 1410. [Google Scholar] [CrossRef] [Green Version]
- Torrino, S.; Visvikis, O.; Doye, A.; Boyer, L.; Stefani, C.; Munro, P.; Bertoglio, J.; Gacon, G.; Mettouchi, A.; Lemichez, E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 2011, 21, 959–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrio, E.; Lotte, R.; Hamaoui, D.; Cherfils, J.; Doye, A.; Daugaard, M.; Sorensen, P.H.; Bost, F.; Ruimy, R.; Mettouchi, A.; et al. Identification of cancer-associated missense mutations in in hace1 that impair cell growth control and Rac1 ubiquitylation. Sci. Rep. 2017, 7, 44779. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Anglesio, M.S.; O’Sullivan, M.; Zhang, F.; Yang, G.; Sarao, R.; Nghiem, M.P.; Cronin, S.; Hara, H.; Melnyk, N.; et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat. Med. 2007, 13, 1060–1069. [Google Scholar] [CrossRef]
- Hibi, K.; Sakata, M.; Sakuraba, K.; Shirahata, A.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008, 28, 1581–1584. [Google Scholar]
- Zhao, J.; Mialki, R.K.; Wei, J.; Coon, T.A.; Zou, C.; Chen, B.B.; Mallampalli, R.K.; Zhao, Y. SCF E3 ligase F-box protein complex SCF FBXL19 regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J. 2013, 27, 2611–2619. [Google Scholar] [CrossRef] [Green Version]
- Oberoi-Khanuja, T.K.; Rajalingam, K. IAPs as E3 ligases of Rac1: Shaping the move. Small GTPases 2012, 3, 131–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberoi, T.K.; Dogan, T.; Hocking, J.C.; Scholz, R.-P.; Mooz, J.; Anderson, C.L.; Karreman, C.; Meyer zu Heringdorf, D.; Schmidt, G.; Ruonala, M.; et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012, 31, 14–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, K.; Fujibuchi, W.; Ishida, K.; Naitoh, T.; Ogawa, H.; Ando, T.; Yazaki, N.; Watanabe, K.; Haneda, S.; Shibata, C.; et al. Inhibitor of apoptosis protein family as diagnostic markers and therapeutic targets of colorectal cancer. Surg. Today 2011, 41, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Voth, D.E.; Ballard, J.D. Clostridium difficile Toxins: Mechanism of Action and Role in Disease. Clin. Microbiol. Rev. 2005, 18, 247–263. [Google Scholar] [CrossRef] [Green Version]
- Yarbrough, M.L.; Li, Y.; Kinch, L.N.; Grishin, N.V.; Ball, H.L.; Orth, K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 2009, 323, 269–272. [Google Scholar] [CrossRef] [Green Version]
- Shao, F.; Dixon, J.E. YopT is a cysteine protease cleaving Rho family GTPases. Adv. Exp. Med. Biol. 2003, 529, 79–84. [Google Scholar]
- Okada, R.; Zhou, X.; Hiyoshi, H.; Matsuda, S.; Chen, X.; Akeda, Y.; Kashimoto, T.; Davis, B.M.; Iida, T.; Waldor, M.K.; et al. The Vibrio parahaemolyticuseffector VopC mediates Cdc42-dependent invasion of cultured cells but is not required for pathogenicity in an animal model of infection. Cell Microbiol. 2014, 16, 938–947. [Google Scholar] [CrossRef] [Green Version]
- Masuda, M.; Betancourt, L.; Matsuzawa, T.; Kashimoto, T.; Takao, T.; Shimonishi, Y.; Horiguchi, Y. Activation of rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000, 19, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Rocha, C.L.; Rucks, E.A.; Vincent, D.M.; Olson, J.C. Examination of the Coordinate Effects of Pseudomonas aeruginosa ExoS on Rac1. Infect. Immun. 2005, 73, 5458–5467. [Google Scholar] [CrossRef] [Green Version]
- Sumigray, K.D.; Terwilliger, M.; Lechler, T. Morphogenesis and Compartmentalization of the Intestinal Crypt. Dev. Cell 2018, 45, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Beaulieu, J.-F. Integrin α6β4 in Colorectal Cancer: Expression, Regulation, Functional Alterations and Use as a Biomarker. Cancers 2020, 12, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chono, E.; Kurokawa, T.; Oda, C.; Kawasaki, K.; Yamamoto, T.; Ishibashi, S. Expression of rac1 protein in the crypt-villus axis of rat small intestine: In reference to insulin action. Biochem. Biophys. Res. Commun. 1997, 233, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Malliri, A.; Rygiel, T.P.; van der Kammen, R.A.; Song, J.-Y.; Engers, R.; Hurlstone, A.F.L.; Clevers, H.; Collard, J.G. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J. Biol. Chem. 2006, 281, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsson, A.-K.E.; Dai, Y.; Nobis, M.; Baker, M.J.; McGhee, E.J.; Walker, S.; Schwarz, J.P.; Kadir, S.; Morton, J.P.; Myant, K.B.; et al. The Rac-FRET Mouse Reveals Tight Spatiotemporal Control of Rac Activity in Primary Cells and Tissues. Cell Rep. 2014, 6, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Carmon, K.S.; Gong, X.; Yi, J.; Wu, L.; Thomas, A.; Moore, C.M.; Masuho, I.; Timson, D.J.; Martemyanov, K.A.; Liu, Q.J. LGR5 receptor promotes cell–cell adhesion in stem cells and colon cancer cells via the IQGAP1–Rac1 pathway. J. Biol. Chem. 2017, 292, 14989–15001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamieson, C.; Lui, C.; Brocardo, M.G.; Martino-Echarri, E.; Henderson, B.R. Rac1 augments Wnt signaling by stimulating β-catenin-lymphoid enhancer factor-1 complex assembly independent of β-catenin nuclear import. J. Cell Sci. 2015, 128, 3933–3946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Myant, K.B.; Cammareri, P.; McGhee, E.J.; Ridgway, R.A.; Huels, D.J.; Cordero, J.B.; Schwitalla, S.; Kalna, G.; Ogg, E.-L.; Athineos, D.; et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 2013, 12, 761–773. [Google Scholar] [CrossRef] [Green Version]
- Myant, K.B.; Scopelliti, A.; Haque, S.; Vidal, M.; Sansom, O.J.; Cordero, J.B. Rac1 drives intestinal stem cell proliferation and regeneration. Cell Cycle 2013, 12, 2973–2977. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.R.; Zeng, X.; Zhao, J.; Liu, Y.; Hou, G.; Liu, H.; Hou, S.X. The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 2016, 538, 109–113. [Google Scholar] [CrossRef]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Stappenbeck, T.S.; Gordon, J.I. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development 2000, 127, 2629–2642. [Google Scholar] [PubMed]
- Wang, S.; Walton, K.D.; Gumucio, D.L. Signals and forces shaping organogenesis of the small intestine. Curr. Top. Dev. Biol. 2019, 132, 31–65. [Google Scholar] [PubMed]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Garcia, M.A.; Nelson, W.J.; Chavez, N. Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb. Perspect Biol. 2018, 10, a029181. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, N.; Meir, M.; Spindler, V.; Germer, C.-T.; Waschke, J. Differential role of Rho GTPases in intestinal epithelial barrier regulation in vitro. J. Cell. Physiol. 2011, 226, 1196–1203. [Google Scholar] [CrossRef]
- Garcia-Cattaneo, A.; Braga, V.M.M. Hold on tightly: How to keep the local activation of small GTPases. Cell Adh. Migr. 2013, 7, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Citi, S.; Guerrera, D.; Spadaro, D.; Shah, J. Epithelial junctions and Rho family GTPases: The zonular signalosome. Small GTPases 2014, 5, e973760. [Google Scholar] [CrossRef] [Green Version]
- Arnold, T.R.; Stephenson, R.E.; Miller, A.L. Rho GTPases and actomyosin: Partners in regulating epithelial cell-cell junction structure and function. Exp. Cell Res. 2017, 358, 20–30. [Google Scholar] [CrossRef]
- Marei, H.; Malliri, A. GEFs: Dual regulation of Rac1 signaling. Small GTPases 2017, 8, 90–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.-B.; Liu, H.-S.; Zhang, L.-J.; Liu, X.-H.; Zhong, X.-L.; Zhou, C.; Hu, T.; Wu, X.-R.; Hu, J.-C.; Lian, L.; et al. Engulfment and Cell Motility Protein 1 Protects Against DSS-induced Colonic Injury in Mice via Rac1 Activation. J. Crohns Colitis 2019, 13, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Leoni, G.; Neumann, P.A.; Chun, J.; Nusrat, A.; Yun, C.C. Distinct Phospholipase C-β Isozymes Mediate Lysophosphatidic Acid Receptor 1 Effects on Intestinal Epithelial Homeostasis and Wound Closure. Mol. Cell. Biol. 2013, 33, 2016–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Qi, Z.; Li, Y.; Wang, C.; Fu, W.; Chen, Y.-G. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015, 6, 7166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidelin, J.B.; Larsen, S.; Linnemann, D.; Vainer, B.; Coskun, M.; Troelsen, J.T.; Nielsen, O.H. Cellular inhibitor of apoptosis protein 2 controls human colonic epithelial restitution, migration, and Rac1 activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G92–G99. [Google Scholar] [CrossRef] [Green Version]
- Birkl, D.; Quiros, M.; Garcia-Hernandez, V.; Zhou, D.W.; Brazil, J.C.; Hilgarth, R.; Keeney, J.; Yulis, M.; Bruewer, M.; García, A.J.; et al. TNFα promotes mucosal wound repair through enhanced platelet activating factor receptor signaling in the epithelium. Mucosal Immunol. 2019, 12, 909–918. [Google Scholar] [CrossRef]
- Power, K.A.; Lu, J.T.; Monk, J.M.; Lepp, D.; Wu, W.; Zhang, C.; Liu, R.; Tsao, R.; Robinson, L.E.; Wood, G.A.; et al. Purified rutin and rutin-rich asparagus attenuates disease severity and tissue damage following dextran sodium sulfate-induced colitis. Mol. Nutr. Food Res. 2016, 60, 2396–2412. [Google Scholar] [CrossRef]
- Monk, J.M.; Wu, W.; Hutchinson, A.L.; Pauls, P.; Robinson, L.E.; Power, K.A. Navy and black bean supplementation attenuates colitis-associated inflammation and colonic epithelial damage. J. Nutr. Biochem. 2018, 56, 215–223. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114. [Google Scholar] [CrossRef]
- den Hartog, G.; Chattopadhyay, R.; Ablack, A.; Hall, E.H.; Butcher, L.D.; Bhattacharyya, A.; Eckmann, L.; Harris, P.R.; Das, S.; Ernst, P.B.; et al. Regulation of Rac1 and Reactive Oxygen Species Production in Response to Infection of Gastrointestinal Epithelia. PLoS Pathog. 2016, 12, e1005382. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.P.; Papaioannou, A.; Malliri, A. Deregulation of Rho GTPases in cancer. Small GTPases 2016, 7, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xu, W.; Lu, T.; Zhou, J.; Ge, X.; Hua, D. MicroRNA-142-3p Promotes Cellular Invasion of Colorectal Cancer Cells by Activation of RAC1. Technol. Cancer Res. Treat. 2018, 17, 153303381879050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Guo, H.; Lu, Y.; Feng, W.; Sun, X.; Tang, C.; Wang, X.; Shen, M. Blocking hepatic metastases of colon cancer cells using an shRNA against Rac1 delivered by activatable cell-penetrating peptide. Oncotarget 2016, 7, 77183–77195. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Lin, J.; Su, J.; Oyang, L.; Wang, H.; Tan, S.; Tang, Y.; Chen, X.; Liu, W.; Luo, X.; et al. Diallyl disulfide inhibits colon cancer metastasis by suppressing Rac1-mediated epithelial-mesenchymal transition. OncoTargets Ther. 2019, 12, 5713–5728. [Google Scholar] [CrossRef] [Green Version]
- Lou, S.; Wang, P.; Yang, J.; Ma, J.; Liu, C.; Zhou, M. Prognostic and Clinicopathological Value of Rac1 in Cancer Survival: Evidence from a Meta-Analysis. J. Cancer 2018, 9, 2571–2579. [Google Scholar] [CrossRef]
- Zhao, H.; Dong, T.; Zhou, H.; Wang, L.; Huang, A.; Feng, B.; Quan, Y.; Jin, R.; Zhang, W.; Sun, J.; et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis 2014, 35, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Zhang, Y.; Lin, S.; Sun, A.; Chen, R.; Ding, Y.; Ding, Y. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10534–10544. [Google Scholar]
- Zhou, K.; Rao, J.; Zhou, Z.-H.; Yao, X.-H.; Wu, F.; Yang, J.; Yang, L.; Zhang, X.; Cui, Y.-H.; Bian, X.-W.; et al. RAC1-GTP promotes epithelial-mesenchymal transition and invasion of colorectal cancer by activation of STAT3. Lab. Investig. 2018, 98, 989–998. [Google Scholar] [CrossRef] [Green Version]
- Payapilly, A.; Malliri, A. Compartmentalisation of RAC1 signalling. Curr. Opin. Cell Biol. 2018, 54, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Hittelman, W.; Lu, T.; Ji, P.; Arlinghaus, R.; Shmulevich, I.; Hamilton, S.R.; Zhang, W. NGAL decreases E-cadherin-mediated cell-cell adhesion and increases cell motility and invasion through Rac1 in colon carcinoma cells. Lab. Investig. 2009, 89, 531–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanning, C.C.; Daddona, J.L.; Ruiz-Velasco, R.; Shafer, S.H.; Williams, C.L. The Rac1 C-terminal Polybasic Region Regulates the Nuclear Localization and Protein Degradation of Rac1. J. Biol. Chem. 2004, 279, 44197–44210. [Google Scholar] [CrossRef] [Green Version]
- Abdrabou, A.; Brandwein, D.; Liu, C.; Wang, Z. Rac1 S71 Mediates the Interaction between Rac1 and 14-3-3 Proteins. Cells 2019, 8, 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, J.; Proff, J.; Hävemeier, A.; Ladwein, M.; Rottner, K.; Barlag, B.; Pich, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS ONE 2012, 7, e44358. [Google Scholar] [CrossRef] [Green Version]
- Schnelzer, A.; Prechtel, D.; Knaus, U.; Dehne, K.; Gerhard, M.; Graeff, H.; Harbeck, N.; Schmitt, M.; Lengyel, E. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000, 19, 3013–3020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.L.; Carmo, F.; Bugalho, M.J. RAC1b overexpression in papillary thyroid carcinoma: A role to unravel. Eur. J. Endocrinol. 2013, 168, 795–804. [Google Scholar] [CrossRef] [Green Version]
- Ungefroren, H.; Sebens, S.; Giehl, K.; Helm, O.; Groth, S.; Fandrich, F.; Rocken, C.; Sipos, B.; Lehnert, H.; Gieseler, F. Rac1b negatively regulates TGF-β1-induced cell motility in pancreatic ductal epithelial cells by suppressing Smad signalling. Oncotarget 2014, 5, 277–290. [Google Scholar] [CrossRef] [Green Version]
- Mehner, C.; Miller, E.; Nassar, A.; Bamlet, W.R.; Radisky, E.S.; Radisky, D.C. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer 2015, 6, 480–489. [Google Scholar]
- Stallings-Mann, M.L.; Waldmann, J.; Zhang, Y.; Miller, E.; Gauthier, M.L.; Visscher, D.W.; Downey, G.P.; Radisky, E.S.; Fields, A.P.; Radisky, D.C. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci. Transl. Med. 2012, 4, 142ra95. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Licciulli, S.; Avila, J.L.; Cho, M.; Troutman, S.; Jiang, P.; Kossenkov, A.V.; Showe, L.C.; Liu, Q.; Vachani, A.; et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2013, 32, 903–909. [Google Scholar] [CrossRef] [Green Version]
- Matos, P.; Kotelevets, L.; Goncalves, V.; Henriques, A.F.A.; Henriques, A.; Zerbib, P.; Moyer, M.P.; Chastre, E.; Jordan, P. Ibuprofen inhibits colitis-induced overexpression of tumor-related Rac1b. Neoplasia 2013, 15, 102–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, V.; Matos, P.; Jordan, P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum. Mol. Genet. 2009, 18, 3696–3707. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.; Henriques, A.F.A.; Henriques, A.; Pereira, J.F.S.; Pereira, J.; Neves Costa, A.; Moyer, M.P.; Moita, L.F.; Gama-Carvalho, M.; Matos, P.; et al. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 2014, 20, 474–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelisch, F.; Khauv, D.; Risso, G.; Stallings-Mann, M.; Blaustein, M.; Quadrana, L.; Radisky, D.C.; Srebrow, A. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J. Cell. Biochem. 2012, 113, 2319–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, H.; Saitoh, M.; Sakamoto, K.; Kondo, T.; Katoh, R.; Tanaka, S.; Motizuki, M.; Masuyama, K.; Miyazawa, K. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem. 2014, 289, 27386–27399. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Fu, X.; Chen, P.; Wu, P.; Fan, X.; Li, N.; Zhu, H.; Jia, T.-T.; Ji, H.; Wang, Z.; et al. SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling. Cell Res. 2017, 27, 540–558. [Google Scholar] [CrossRef] [Green Version]
- Matos, P.; Collard, J.G.; Jordan, P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J. Biol. Chem. 2003, 278, 50442–50448. [Google Scholar] [CrossRef] [Green Version]
- Matos, P.; Jordan, P. Increased Rac1b expression sustains colorectal tumor cell survival. Mol. Cancer Res. 2008, 6, 1178–1184. [Google Scholar] [CrossRef] [Green Version]
- Matos, P.; Jordan, P. Rac1, but not Rac1B, stimulates RelB-mediated gene transcription in colorectal cancer cells. J. Biol. Chem. 2006, 281, 13724–13732. [Google Scholar] [CrossRef] [Green Version]
- Lozano, E.; Frasa, M.A.M.; Smolarczyk, K.; Knaus, U.G.; Braga, V.M.M. PAK is required for the disruption of E-cadherin adhesion by the small GTPase Rac. J. Cell Sci. 2008, 121, 933–938. [Google Scholar] [CrossRef] [Green Version]
- Orlichenko, L.; Geyer, R.; Yanagisawa, M.; Khauv, D.; Radisky, E.S.; Anastasiadis, P.Z.; Radisky, D.C. The 19-amino acid insertion in the tumor-associated splice isoform Rac1b confers specific binding to p120 catenin. J. Biol. Chem. 2010, 285, 19153–19161. [Google Scholar] [CrossRef] [Green Version]
- Esufali, S.; Charames, G.S.; Pethe, V.V.; Buongiorno, P.; Bapat, B. Activation of tumor-specific splice variant Rac1b by dishevelled promotes canonical Wnt signaling and decreased adhesion of colorectal cancer cells. Cancer Res. 2007, 67, 2469–2479. [Google Scholar] [CrossRef] [Green Version]
- Radisky, D.C.; Levy, D.D.; Littlepage, L.E.; Liu, H.; Nelson, C.M.; Fata, J.E.; Leake, D.; Godden, E.L.; Albertson, D.G.; Angela Nieto, M.; et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005, 436, 123–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Chen, Q.K.; Lui, C.; Cichon, M.A.; Radisky, D.C.; Nelson, C.M. Matrix compliance regulates Rac1b localization, NADPH oxidase assembly, and epithelial-mesenchymal transition. Mol. Biol. Cell 2012, 23, 4097–4108. [Google Scholar] [CrossRef] [PubMed]
- Witte, D.; Otterbein, H.; Rster, M.F.X.; Giehl, K.; Zeiser, R.; Lehnert, H.; Ungefroren, H. Negative regulation of TGF-β1-induced MKK6-p38 and MEK-ERK signalling and epithelial-mesenchymal transition by Rac1b. Sci. Rep. 2017, 7, 17313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; Hass, R.; Ohe von der, J.; Lehnert, H.; Ungefroren, H. The role of TGF-β and its crosstalk with RAC1/RAC1b signaling in breast and pancreas carcinoma. Cell Commun. Signal. 2017, 15, 19. [Google Scholar] [CrossRef] [Green Version]
- Matos, P.; Jordan, P. Expression of Rac1b stimulates NF-kappaB-mediated cell survival and G1/S progression. Exp. Cell Res. 2005, 305, 292–299. [Google Scholar] [CrossRef]
- Singh, A.; Karnoub, A.E.; Palmby, T.R.; Lengyel, E.; Sondek, J.; Der, C.J. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene 2004, 23, 9369–9380. [Google Scholar] [CrossRef] [Green Version]
- Henriques, A.F.A.; Barros, P.; Moyer, M.P.; Matos, P.; Jordan, P. Expression of tumor-related Rac1b antagonizes B-Raf-induced senescence in colorectal cells. Cancer Lett. 2015, 369, 368–375. [Google Scholar] [CrossRef]
- Matos, P.; Oliveira, C.; Velho, S.; Goncalves, V.; da Costa, L.T.; Moyer, M.P.; Seruca, R.; Jordan, P. B-Raf(V600E) cooperates with alternative spliced Rac1b to sustain colorectal cancer cell survival. Gastroenterology 2008, 135, 899–906. [Google Scholar] [CrossRef]
- Alonso-Espinaco, V.; Cuatrecasas, M.; Alonso, V.; Escudero, P.; Marmol, M.; Horndler, C.; Ortego, J.; Gallego, R.; Codony-Servat, J.; Garcia-Albeniz, X.; et al. RAC1b overexpression correlates with poor prognosis in KRAS/BRAF WT metastatic colorectal cancer patients treated with first-line FOLFOX/XELOX chemotherapy. Eur. J. Cancer 2014, 50, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.T.; Chaturvedi, P.; Lopez, D.T.M.; Garza, A.D.L.; Lippman, M.E. RAC1b Overexpression Confers Resistance to Chemotherapy Treatment in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Chernoff, J.; Zheng, Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J. Biol. Chem. 1998, 273, 8776–8782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, S.; Deng, Y.; Fu, J.; Zhang, Y.; Zhang, Z.; Ru, X.; Qin, X. Decreased expression of ARHGAP15 promotes the development of colorectal cancer through PTEN/AKT/FOXO1 axis. Cell Death Dis. 2018, 9, 673. [Google Scholar] [CrossRef]
- Akilesh, S.; Suleiman, H.; Yu, H.; Stander, M.C.; Lavin, P.; Gbadegesin, R.; Antignac, C.; Pollak, M.; Kopp, J.B.; Winn, M.P.; et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J. Clin. Investig. 2011, 121, 4127–4137. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Sui, L.; Zhuang, J.; He, S.; Song, Y.; Ye, Y.; Xia, W. ARHGAP24 regulates cell ability and apoptosis of colorectal cancer cells via the regulation of P53. Oncol. Lett. 2018, 16, 3517–3524. [Google Scholar]
- Nakamura, T.; Komiya, M.; Sone, K.; Hirose, E.; Gotoh, N.; Morii, H.; Ohta, Y.; Mori, N. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 2002, 22, 8721–8734. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Ma, H.; Bossy-Wetzel, E.; Lipton, S.A.; Zhang, Z.; Feng, G.-S. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J. Biol. Chem. 2003, 278, 34641–34653. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.Y.; Zang, H.; Zheng, Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J. Biol. Chem. 2003, 278, 4151–4159. [Google Scholar] [CrossRef] [Green Version]
- Ge, W.; Cai, W.; Bai, R.; Hu, W.; Wu, D.; Zheng, S.; Hu, H. A novel 4-gene prognostic signature for hypermutated colorectal cancer. Cancer Manag. Res. 2019, 11, 1985–1996. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, M.B.; Rossing, M.; Østrup, O.; Larsen, P.N.; Heiberg Engel, P.J.; Jørgensen, L.N.; Hogdall, E.V.; Eriksen, J.; Ibsen, P.; Jess, P.; et al. Genomic alterations accompanying tumour evolution in colorectal cancer: Tracking the differences between primary tumours and synchronous liver metastases by whole-exome sequencing. BMC Cancer 2018, 18, 752. [Google Scholar] [CrossRef]
- Parrini, M.C.; Sadou-Dubourgnoux, A.; Aoki, K.; Kunida, K.; Biondini, M.; Hatzoglou, A.; Poullet, P.; Formstecher, E.; Yeaman, C.; Matsuda, M.; et al. SH3BP1, an exocyst-associated RhoGAP, inactivates Rac1 at the front to drive cell motility. Mol. Cell 2011, 42, 650–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Feng, Y.; Chen, X.; Du, Z.; Jiang, S.; Ma, S.; Zou, W. SH3BP1-induced Rac-Wave2 pathway activation regulates cervical cancer cell migration, invasion, and chemoresistance to cisplatin. J. Cell. Biochem. 2018, 119, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Zago, G.; Biondini, M.; Camonis, J.; Parrini, M.C. A family affair: A Ral-exocyst-centered network links Ras, Rac, Rho signaling to control cell migration. Small GTPases 2019, 10, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, H.; Toiyama, Y.; Saigusa, S.; Kawamura, M.; Kawamoto, A.; Okugawa, Y.; Hiro, J.; Tanaka, K.; Inoue, Y.; Mohri, Y.; et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2015, 36, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.-M.; Sung, W.-W.; Lai, H.-W.; Hsieh, M.-J.; Yen, H.-H.; Su, T.-C.; Chang, W.-H.; Chen, C.-Y.; Ko, J.-L.; Chen, C.-J. Opposing prognostic roles of nuclear and cytoplasmic RACGAP1 expression in colorectal cancer patients. Hum. Pathol. 2016, 47, 45–51. [Google Scholar] [CrossRef]
- Lian, L.Y.; Barsukov, I.; Golovanov, A.P.; Hawkins, D.I.; Badii, R.; Sze, K.H.; Keep, N.H.; Bokoch, G.M.; Roberts, G.C. Mapping the binding site for the GTP-binding protein Rac-1 on its inhibitor RhoGDI-1. Structure 2000, 8, 47–55. [Google Scholar] [CrossRef]
- Fauré, J.; Dagher, M.C. Interactions between Rho GTPases and Rho GDP dissociation inhibitor (Rho-GDI). Biochimie 2001, 83, 409–414. [Google Scholar] [CrossRef]
- Del Pozo, M.A.; Kiosses, W.B.; Alderson, N.B.; Meller, N.; Hahn, K.M.; Schwartz, M.A. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 2002, 4, 232–239. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, J.-T.; Baek, K.E.; Kim, B.-Y.; Lee, H.G. Regulation of Rho GTPases by RhoGDIs in Human Cancers. Cells 2019, 8, 1037. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Lu, W.; Zou, S.; Wang, H.; Jiang, Y.; Zhang, X.; Li, P.; Songyang, Z.; Wang, L.; Wang, J.; et al. Rho GDP-dissociation inhibitor α is a potential prognostic biomarker and controls telomere regulation in colorectal cancer. Cancer Sci. 2017, 108, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Baird, D.; Cerione, R.A. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 2004, 23, 3492–3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zheng, T. Screening of Hub Genes and Pathways in Colorectal Cancer with Microarray Technology. Pathol. Oncol. Res. 2014, 20, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Klooster ten, J.P.; Jaffer, Z.M.; Chernoff, J.; Hordijk, P.L. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 2006, 172, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Deng, L.; Liu, D.; Liao, S.; Dai, H.; Li, J.; Rong, J.; Wang, Z.; Huang, G.; Tang, C.; et al. ARHGEF7 promotes metastasis of colorectal adenocarcinoma by regulating the motility of cancer cells. Int. J. Oncol. 2018, 53, 1980–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishaque, N.; Abba, M.L.; Hauser, C.; Patil, N.; Paramasivam, N.; Huebschmann, D.; Leupold, J.H.; Balasubramanian, G.P.; Kleinheinz, K.; Toprak, U.H.; et al. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat. Commun. 2018, 9, 4782. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-X.; Zheng, H.-T.; Peng, J.-J.; Huang, L.-Y.; Shi, D.-B.; Liang, L.; Cai, S.-J. RNA-seq reveals determinants for irinotecan sensitivity/resistance in colorectal cancer cell lines. Int. J. Clin. Exp. Pathol. 2014, 7, 2729–2736. [Google Scholar]
- Kawasaki, Y.; Sato, R.; Akiyama, T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat. Cell Biol. 2003, 5, 211–215. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Tsuji, S.; Muroya, K.; Furukawa, S.; Shibata, Y.; Okuno, M.; Ohwada, S.; Akiyama, T. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009, 10, 1355–1362. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, Y.; Furukawa, S.; Sato, R.; Akiyama, T. Differences in the localization of the adenomatous polyposis coli-Asef/Asef2 complex between adenomatous polyposis coli wild-type and mutant cells. Cancer Sci. 2013, 104, 1135–1138. [Google Scholar] [CrossRef]
- Komander, D.; Patel, M.; Laurin, M.; Fradet, N.; Pelletier, A.; Barford, D.; Côté, J.-F. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 2008, 19, 4837–4851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiyokawa, E.; Hashimoto, Y.; Kobayashi, S.; Sugimura, H.; Kurata, T.; Matsuda, M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998, 12, 3331–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 2001, 107, 27–41. [Google Scholar] [CrossRef] [Green Version]
- Makino, Y.; Tsuda, M.; Ichihara, S.; Watanabe, T.; Sakai, M.; Sawa, H.; Nagashima, K.; Hatakeyama, S.; Tanaka, S. Elmo1 inhibits ubiquitylation of Dock180. J. Cell Sci. 2006, 119, 923–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, D.T.; McShea, K.M.; Attar, M.A.; Santy, L.C. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol. Biol. Cell 2010, 21, 562–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, M.; Chiang, T.-C.; Tran, V.; Lee, F.-J.S.; Côté, J.-F. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 2011, 286, 38969–38979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Li, Y.; Yin, Y.; Zhang, W.; Hou, Y.; Zhang, L.; Li, Z.; Xie, B.; Gao, W.-Q.; Sarkaria, J.N.; et al. Protein kinase A-dependent phosphorylation of Dock180 at serine residue 1250 is important for glioma growth and invasion stimulated by platelet derived-growth factor receptor α. NeuroOncology 2015, 17, 832–842. [Google Scholar] [CrossRef] [Green Version]
- Brugnera, E.; Haney, L.; Grimsley, C.; Lu, M.; Walk, S.F.; Tosello-Trampont, A.-C.; Macara, I.G.; Madhani, H.; Fink, G.R.; Ravichandran, K.S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002, 4, 574–582. [Google Scholar] [CrossRef]
- Lu, M.; Kinchen, J.M.; Rossman, K.L.; Grimsley, C.; Hall, M.; Sondek, J.; Hengartner, M.O.; Yajnik, V.; Ravichandran, K.S. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005, 15, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Jing, X.; Wu, H.; Ji, X.; Wu, H.; Shi, M.; Zhao, R. Cortactin promotes cell migration and invasion through upregulation of the dedicator of cytokinesis 1 expression in human colorectal cancer. Oncol. Rep. 2016, 36, 1946–1952. [Google Scholar] [CrossRef]
- Hirakawa, H.; Shibata, K.; Nakayama, T. Localization of cortactin is associated with colorectal cancer development. Int. J. Oncol. 2009, 35, 1271–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, H.; Kobayashi, S.; Hashimoto, Y.; Ohba, F.; Mochizuki, N.; Kurata, T.; Nagashima, K.; Matsuda, M. Non-adherent cell-specific expression of DOCK2, a member of the human CDM-family proteins. Biochim. Biophys. Acta 1999, 1452, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Miao, S.; Zhang, R.-Y.; Wang, W.; Wang, H.-B.; Meng, L.-L.; Zu, L.-D.; Fu, G.-H. Overexpression of dedicator of cytokinesis 2 correlates with good prognosis in colorectal cancer associated with more prominent CD8+ lymphocytes infiltration: A colorectal cancer analysis. J. Cell. Biochem. 2018, 119, 8962–8970. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, W.K.K.; Li, X.; He, J.; Li, X.-X.; Ng, S.S.M.; Yu, C.; Gao, Z.; Yang, J.; Li, M.; et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2015, 64, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Namekata, K.; Enokido, Y.; Iwasawa, K.; Kimura, H. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 2004, 279, 14331–14337. [Google Scholar] [CrossRef] [Green Version]
- Hofer, P.; Hagmann, M.; Brezina, S.; Dolejsi, E.; Mach, K.; Leeb, G.; Baierl, A.; Buch, S.; Sutterlüty-Fall, H.; Karner-Hanusch, J.; et al. Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas. Oncotarget 2017, 8, 98623–98634. [Google Scholar] [CrossRef]
- Upadhyay, G.; Goessling, W.; North, T.E.; Xavier, R.; Zon, L.I.; Yajnik, V. Molecular association between β-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/β-catenin signaling. Oncogene 2008, 27, 5845–5855. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Zhou, J.; Tong, Y. Gene signatures of drug resistance predict patient survival in colorectal cancer. Pharmacogenomics 2015, 15, 135–143. [Google Scholar] [CrossRef]
- Ashraf, S.Q.; Nicholls, A.M.; Wilding, J.L.; Ntouroupi, T.G.; Mortensen, N.J.; Bodmer, W.F. Direct and immune mediated antibody targeting of ERBB receptors in a colorectal cancer cell-line panel. Proc. Natl. Acad. Sci. USA 2012, 109, 21046–21051. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, J.; Miyamoto, Y.; Chan, J.R.; Tanoue, A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell Biol. 2008, 181, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Watabe-Uchida, M.; John, K.A.; Janas, J.A.; Newey, S.E.; Van Aelst, L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron 2006, 51, 727–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arandjelovic, S.; Perry, J.S.A.; Lucas, C.D.; Penberthy, K.K.; Kim, T.-H.; Zhou, M.; Rosen, D.A.; Chuang, T.-Y.; Bettina, A.M.; Shankman, L.S.; et al. A noncanonical role for the engulfment gene ELMO1 in neutrophils that promotes inflammatory arthritis. Nat. Immunol. 2019, 20, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Libanje, F.; Raingeaud, J.; Luan, R.; Thomas, Z.; Zajac, O.; Veiga, J.; Marisa, L.; Adam, J.; Boige, V.; Malka, D.; et al. ROCK2 inhibition triggers the collective invasion of colorectal adenocarcinomas. EMBO J. 2019, 38, e99299. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.R.; Johnson, R.C.; Darlington, D.N.; Hand, T.A.; Mains, R.E.; Eipper, B.A. Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine alpha-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 1997, 272, 12667–12675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabiner, C.A.; Mains, R.E.; Eipper, B.A. Kalirin: A dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist 2005, 11, 148–160. [Google Scholar] [CrossRef]
- Wu, J.-H.; Fanaroff, A.C.; Sharma, K.C.; Smith, L.S.; Brian, L.; Eipper, B.A.; Mains, R.E.; Freedman, N.J.; Zhang, L. Kalirin promotes neointimal hyperplasia by activating Rac in smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 702–708. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.; Wu, J.; Huang, Q.; Yang, X.; Yang, K.; Qiu, Y.; Ren, J.; Shen, R.; Qi, H. NEDD9 promotes invasion and migration of colorectal cancer cell line HCT116 via JNK/EMT. Oncol. Lett. 2019, 18, 4022–4029. [Google Scholar] [CrossRef]
- Dai, J.; Van Wie, P.G.; Fai, L.Y.; Kim, D.; Wang, L.; Poyil, P.; Luo, J.; Zhang, Z. Downregulation of NEDD9 by apigenin suppresses migration, invasion, and metastasis of colorectal cancer cells. Toxicol. Appl. Pharmacol. 2016, 311, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bavarva, J.H.; Wang, Z.; Guo, J.; Qian, C.; Thibodeau, S.N.; Golemis, E.A.; Liu, W. HEF1, a novel target of Wnt signaling, promotes colonic cell migration and cancer progression. Oncogene 2011, 30, 2633–2643. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhou, H.; Zhu, X.; Ma, G.; Liu, C.; Lin, B.; Mao, W. High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients. Int. J. Clin. Exp. Pathol. 2014, 7, 2565–2570. [Google Scholar]
- Barrows, D.; He, J.Z.; Parsons, R. PREX1 Protein Function Is Negatively Regulated Downstream of Receptor Tyrosine Kinase Activation by p21-activated Kinases (PAKs). J. Biol. Chem. 2016, 291, 20042–20054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cash, J.N.; Davis, E.M.; Tesmer, J.J.G. Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3. Structure 2016, 24, 730–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Wang, Q.; Wang, Y.; Chen, X.; Wang, Z. PLC-gamma1 and Rac1 coregulate EGF-induced cytoskeleton remodeling and cell migration. Mol. Endocrinol. 2009, 23, 901–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, R.C.; Cowell, C.A.M.; Hammond, C.L.; Bergen, D.J.M.; Roper, J.A.; Feng, Y.; Rendall, T.C.S.; Race, P.R.; Bass, M.D. Coronin-1C and RCC2 guide mesenchymal migration by trafficking Rac1 and controlling GEF exposure. J. Cell Sci. 2014, 127, 4292–4307. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Liang, L.; Jin, Y.; Li, Y.; Liu, Y.; Guo, L.; Wu, C.; Yun, C.-H.; Yin, Y. RCC2 is a novel p53 target in suppressing metastasis. Oncogene 2018, 37, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Bruun, J.; Kolberg, M.; Ahlquist, T.C.; Røyrvik, E.C.; Nome, T.; Leithe, E.; Lind, G.E.; Merok, M.A.; Rognum, T.O.; Bjørkøy, G.; et al. Regulator of Chromosome Condensation 2 Identifies High-Risk Patients within Both Major Phenotypes of Colorectal Cancer. Clin. Cancer Res. 2015, 21, 3759–3770. [Google Scholar] [CrossRef] [Green Version]
- Nimnual, A.S.; Yatsula, B.A.; Bar-Sagi, D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 1998, 279, 560–563. [Google Scholar] [CrossRef]
- Malliri, A.; van Es, S.; Huveneers, S.; Collard, J.G. The Rac Exchange Factor Tiam1 Is Required for the Establishment and Maintenance of Cadherin-based Adhesions. J. Biol. Chem. 2004, 279, 30092–30098. [Google Scholar] [CrossRef] [Green Version]
- Michiels, F.; Habets, G.G.; Stam, J.C.; van der Kammen, R.A.; Collard, J.G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature 1995, 375, 338–340. [Google Scholar] [CrossRef]
- Ehler, E.; van Leeuwen, F.; Collard, J.G.; Salinas, P.C. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol. Cell. Neurosci. 1997, 9, 1–12. [Google Scholar] [CrossRef]
- Liu, L.; Wu, D.-H.; Ding, Y.-Q. Tiam1 gene expression and its significance in colorectal carcinoma. World J. Gastroenterol. 2005, 11, 705–707. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Q.; Zhang, Y.; Wang, S.; Ding, Y. Lentivirus-Mediated Silencing of Tiam1 Gene Influences Multiple Functions of a Human Colorectal Cancer Cell Line. Neoplasia 2006, 8, 917–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Li, W.; Liu, H.; Yang, L.; Liao, Q.; Cui, S.; Wang, H.; Zhao, L. miR-29b suppresses tumor growth and metastasis in colorectal cancer via downregulating Tiam1 expression and inhibiting epithelial-mesenchymal transition. Cell Death Dis. 2014, 5, e1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.-H.; Li, Y.-G.; Han, Z.-P.; Zhou, J.-B.; Chen, W.-M.; Lv, Y.-B.; He, M.-L.; Zuo, J.-D.; Zheng, L. The CircRNA-ACAP2/Hsa-miR-21-5p/ Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells. Cell Physiol. Biochem. 2018, 49, 1539–1550. [Google Scholar] [CrossRef]
- Yu, L.-N.; Zhang, Q.-L.; Li, X.; Hua, X.; Cui, Y.-M.; Zhang, N.-J.; Liao, W.-T.; Ding, Y.-Q. Tiam1 Transgenic Mice Display Increased Tumor Invasive and Metastatic Potential of Colorectal Cancer after 1,2-Dimethylhydrazine Treatment. PLoS ONE 2013, 8, e73077. [Google Scholar] [CrossRef]
- Minard, M.E.; Ellis, L.M.; Gallick, G.E. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin. Exp. Metastasis 2006, 23, 301–313. [Google Scholar] [CrossRef]
- Izumi, D.; Toden, S.; Ureta, E.; Ishimoto, T.; Baba, H.; Goel, A. TIAM1 promotes chemoresistance and tumor invasiveness in colorectal cancer. Cell Death Dis. 2019, 10, 267. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Fan, Z.; Ding, M.; Zhang, H.; Mu, L.; Ding, Y.; Zhang, Y.; Jia, B.; Chen, L.; Chang, Z.; et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene 2015, 34, 5971–5982. [Google Scholar] [CrossRef]
- Hornstein, I.; Alcover, A.; Katzav, S. Vav proteins, masters of the world of cytoskeleton organization. Cell. Signal. 2004, 16, 1–11. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Wang, J.-Y.; Chang, H.-J.; Kuo, C.-W.; Tok, T.-S.; Lin, S.-R. CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol. Rep. 2011, 25, 1297–1306. [Google Scholar]
- Sho, S.; Court, C.M.; Winograd, P.; Russell, M.M.; Tomlinson, J.S. A prognostic mutation panel for predicting cancer recurrence in stages II and III colorectal cancer. J. Surg. Oncol. 2017, 116, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Kroviarski, Y.; Debbabi, M.; Bachoual, R.; Périanin, A.; Gougerot-Pocidalo, M.-A.; El-Benna, J.; Dang, P.M.-C. Phosphorylation of NADPH oxidase activator 1 (NOXA1) on serine 282 by MAP kinases and on serine 172 by protein kinase C and protein kinase A prevents NOX1 hyperactivation. FASEB J. 2010, 24, 2077–2092. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, D.; Abo, A.; Johnston, C.; Segal, A.W.; Hall, A. Interaction of Rac with p67phox and regulation of phagocytic NADPH oxidase activity. Science 1994, 265, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Gorzalczany, Y.; Alloul, N.; Sigal, N.; Weinbaum, C.; Pick, E. A prenylated p67phox-Rac1 chimera elicits NADPH-dependent superoxide production by phagocyte membranes in the absence of an activator and of p47phox: Conversion of a pagan NADPH oxidase to monotheism. J. Biol. Chem. 2002, 277, 18605–18610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.; Prigmore, E.; Govind, S.; Veryard, C.; Kozma, R.; Wientjes, F.B.; Segal, A.W.; Lim, L. Cryptic Rac-binding and p21(Cdc42Hs/Rac)-activated kinase phosphorylation sites of NADPH oxidase component p67(phox). J. Biol. Chem. 1998, 273, 15693–15701. [Google Scholar] [CrossRef] [Green Version]
- Prigmore, E.; Ahmed, S.; Best, A.; Kozma, R.; Manser, E.; Segal, A.W.; Lim, L. A 68-kDa kinase and NADPH oxidase component p67phox are targets for Cdc42Hs and Rac1 in neutrophils. J. Biol. Chem. 1995, 270, 10717–10722. [Google Scholar] [CrossRef] [Green Version]
- Lapouge, K.; Smith, S.J.; Walker, P.A.; Gamblin, S.J.; Smerdon, S.J.; Rittinger, K. Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 2000, 6, 899–907. [Google Scholar] [CrossRef]
- Rolland, T.; Taşan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, N.; Xia, P.; Wang, E.; Guo, Q.; Ye, Z. Inhibition of Rac1 ameliorates neuronal oxidative stress damage via reducing Bcl-2/Rac1 complex formation in mitochondria through PI3K/Akt/mTOR pathway. Exp. Neurol. 2018, 300, 149–166. [Google Scholar] [CrossRef]
- Huang, Q.; Li, S.; Cheng, P.; Deng, M.; He, X.; Wang, Z.; Yang, C.-H.; Zhao, X.-Y.; Huang, J. High expression of anti-apoptotic protein Bcl-2 is a good prognostic factor in colorectal cancer: Result of a meta-analysis. World J. Gastroenterol. 2017, 23, 5018–5033. [Google Scholar] [CrossRef]
- Osborn-Heaford, H.L.; Ryan, A.J.; Murthy, S.; Racila, A.-M.; He, C.; Sieren, J.C.; Spitz, D.R.; Carter, A.B. Mitochondrial Rac1 GTPase import and electron transfer from cytochrome c are required for pulmonary fibrosis. J. Biol. Chem. 2012, 287, 3301–3312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuncewicz, T.; Balakrishnan, P.; Snuggs, M.B.; Kone, B.C. Specific association of nitric oxide synthase-2 with Rac isoforms in activated murine macrophages. Am. J. Physiol. Renal Physiol. 2001, 281, F326–F336. [Google Scholar] [CrossRef] [PubMed]
- Buongiorno, P.; Pethe, V.V.; Charames, G.S.; Esufali, S.; Bapat, B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol. Cancer 2008, 7, 73. [Google Scholar] [CrossRef] [Green Version]
- Esufali, S.; Bapat, B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of β-catenin and TCF/LEF-mediated transcriptional activation. Oncogene 2004, 23, 8260–8271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, A.R.; Vikis, H.G.; Stewart, S.; Fanburg, B.L.; Cochran, B.H.; Guan, K.L. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science 2000, 290, 144–147. [Google Scholar] [CrossRef]
- Zhang, F.; Lu, Y.-X.; Chen, Q.; Zou, H.-M.; Zhang, J.-M.; Hu, Y.-H.; Li, X.-M.; Zhang, W.-J.; Zhang, W.; Lin, C.; et al. Identification of NCK1 as a novel downstream effector of STAT3 in colorectal cancer metastasis and angiogenesis. Cell. Signal. 2017, 36, 67–78. [Google Scholar] [CrossRef]
- Lorès, P.; Visvikis, O.; Luna, R.; Lemichez, E.; Gacon, G. The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J. 2010, 277, 1453–1464. [Google Scholar] [CrossRef]
- Pérez-Yépez, E.A.; Saldívar-Cerón, H.I.; Villamar-Cruz, O.; Pérez-Plasencia, C.; Arias-Romero, L.E. p21 Activated kinase 1: Nuclear activity and its role during DNA damage repair. DNA Repair 2018, 65, 42–46. [Google Scholar] [CrossRef]
- Rane, C.K.; Minden, A. P21 activated kinase signaling in cancer. Semin. Cancer Biol. 2019, 54, 40–49. [Google Scholar] [CrossRef]
- Song, B.; Wang, W.; Zheng, Y.; Yang, J.; Xu, Z. P21-activated kinase 1 and 4 were associated with colorectal cancer metastasis and infiltration. J. Surg. Res. 2015, 196, 130–135. [Google Scholar] [CrossRef]
- Wang, S.E.; Shin, I.; Wu, F.Y.; Friedman, D.B.; Arteaga, C.L. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor beta. Cancer Res. 2006, 66, 9591–9600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, H.; Nicholas, N.S.; Wells, C.M. Role of p-21-activated kinases in cancer progression. Int. Rev. Cell Mol. Biol. 2014, 309, 347–387. [Google Scholar] [PubMed]
- Li, Y.; Shao, Y.; Tong, Y.; Shen, T.; Zhang, J.; Li, Y.; Gu, H.; Li, F. Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 465–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, W.; An, Z.; Wang, Y.; Pan, X.; Fang, W.; Jiang, B.; Zhang, H. P21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. Int. J. Cancer 2009, 125, 548–555. [Google Scholar] [CrossRef]
- Fanger, G.R.; Johnson, N.L.; Johnson, G.L. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997, 16, 4961–4972. [Google Scholar] [CrossRef] [Green Version]
- Nagata, K.I.; Puls, A.; Futter, C.; Aspenstrom, P.; Schaefer, E.; Nakata, T.; Hirokawa, N.; Hall, A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998, 17, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Gerwins, P.; Blank, J.L.; Johnson, G.L. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J. Biol. Chem. 1997, 272, 8288–8295. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, S.; Fukata, M.; Kobayashi, K.; Nakafuku, M.; Nomura, N.; Iwamatsu, A.; Kaibuchi, K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 1996, 271, 23363–23367. [Google Scholar] [CrossRef] [Green Version]
- Hart, M.J.; Callow, M.G.; Souza, B.; Polakis, P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996, 15, 2997–3005. [Google Scholar] [CrossRef]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef] [Green Version]
- Nabeshima, K.; Shimao, Y.; Inoue, T.; Koono, M. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: Its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002, 176, 101–109. [Google Scholar] [CrossRef]
- Brill, S.; Li, S.; Lyman, C.W.; Church, D.M.; Wasmuth, J.J.; Weissbach, L.; Bernards, A.; Snijders, A.J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996, 16, 4869–4878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Hassan, M.K.; Pattnaik, N.; Mohapatra, N.; Dixit, M. Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers. PLoS ONE 2017, 12, e0186977. [Google Scholar] [CrossRef] [PubMed]
- Katkoori, V.R.; Shanmugam, C.; Jia, X.; Vitta, S.P.; Sthanam, M.; Callens, T.; Messiaen, L.; Chen, D.; Zhang, B.; Bumpers, H.L.; et al. Prognostic Significance and Gene Expression Profiles of p53 Mutations in Microsatellite-Stable Stage III Colorectal Adenocarcinomas. PLoS ONE 2012, 7, e30020. [Google Scholar] [CrossRef] [Green Version]
- Tapon, N.; Nagata, K.; Lamarche, N.; Hall, A. A new rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kappaB signalling pathways. EMBO J. 1998, 17, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Kukekov, N.V.; Greene, L.A. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 2003, 22, 252–261. [Google Scholar] [CrossRef] [Green Version]
- Kärkkäinen, S.; van der Linden, M.; Renkema, G.H. POSH2 is a RING finger E3 ligase with Rac1 binding activity through a partial CRIB domain. FEBS Lett. 2010, 584, 3867–3872. [Google Scholar] [CrossRef] [Green Version]
- Uhlik, M.T.; Abell, A.N.; Johnson, N.L.; Sun, W.; Cuevas, B.D.; Lobel-Rice, K.E.; Horne, E.A.; Dell’Acqua, M.L.; Johnson, G.L. Rac–MEKK3–MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 2003, 5, 1104–1110. [Google Scholar] [CrossRef]
- Sandrock, K.; Bielek, H.; Schradi, K.; Schmidt, G.; Klugbauer, N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 2010, 11, 198–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Yu, F.; Lu, S.; Sun, H.; Tang, H.; Peng, Z. Karyopherin alpha 2 is a novel prognostic marker and a potential therapeutic target for colon cancer. J. Exp. Clin. Cancer Res. 2015, 34, 145. [Google Scholar] [CrossRef] [Green Version]
- Perathoner, A.; Pirkebner, D.; Brandacher, G.; Spizzo, G.; Stadlmann, S.; Obrist, P.; Margreiter, R.; Amberger, A. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 2005, 11, 3274–3279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamel, B.; Monaghan-Benson, E.; Rojas, R.J.; Temple, B.R.S.; Marston, D.J.; Burridge, K.; Sondek, J. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J. Biol. Chem. 2011, 286, 12141–12148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.-I.; Fukumoto, Y.; Nochioka, K.; Minami, T.; Kudo, S.; Shiba, N.; Takai, Y.; Williams, C.L.; Liao, J.K.; Shimokawa, H. Statins exert the pleiotropic effects through small GTP-binding protein dissociation stimulator upregulation with a resultant Rac1 degradation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1591–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, T.J.; Gastonguay, A.J.; Lorimer, E.L.; Kuhnmuench, J.R.; Li, R.; Fields, A.P.; Williams, C.L. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 2010, 285, 35255–35266. [Google Scholar] [CrossRef] [Green Version]
- Van Aelst, L.; Joneson, T.; Bar-Sagi, D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996, 15, 3778–3786. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Boshans, R.L.; McDonough, M.; Stahl, P.D.; Van Aelst, L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997, 16, 5445–5454. [Google Scholar] [CrossRef] [Green Version]
- Shin, O.H.; Exton, J.H. Differential binding of arfaptin 2/POR1 to ADP-ribosylation factors and Rac1. Biochem. Biophys. Res. Commun. 2001, 285, 1267–1273. [Google Scholar] [CrossRef]
- Miki, H.; Yamaguchi, H.; Suetsugu, S.; Takenawa, T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 2000, 408, 732–735. [Google Scholar] [CrossRef]
- Williamson, R.C.; Cowell, C.A.M.; Reville, T.; Roper, J.A.; Rendall, T.C.S.; Bass, M.D. Coronin-1C Protein and Caveolin Protein Provide Constitutive and Inducible Mechanisms of Rac1 Protein Trafficking. J. Biol. Chem. 2015, 290, 15437–15449. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Kuroda, S.; Fukata, M.; Nakamura, T.; Nagase, T.; Nomura, N.; Matsuura, Y.; Yoshida-Kubomura, N.; Iwamatsu, A.; Kaibuchi, K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 1998, 273, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.; Qin, H.; Bahassan, A.; Bendzunas, N.G.; Kennedy, E.J.; Cowell, J.K. The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res. 2016, 76, 5133–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shifrin, Y.; Pinto, V.I.; Hassanali, A.; Arora, P.D.; McCulloch, C.A. Force-induced apoptosis mediated by the Rac/Pak/p38 signalling pathway is regulated by filamin A. Biochem. J. 2012, 445, 57–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Z.-Q.; Shi, J.-W.; Wang, X.-R.; Li, Z.; Wang, G.-Y. New cancer suppressor gene for colorectal adenocarcinoma: Filamin A. World J. Gastroenterol. 2015, 21, 2199–2205. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Ka, S.H.; Cho, Y.; Choi, E.-J.; Baek, S.H.; Seol, J.H.; Park, D.; et al. Filamin B serves as a molecular scaffold for type I interferon-induced c-Jun NH2-terminal kinase signaling pathway. Mol. Biol. Cell 2008, 19, 5116–5130. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Kim, S.M.; Ka, S.H.; Oh, K.H.; Kim, I.K.; Zhang, D.-E.; Bang, O.S.; et al. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009, 10, 374–380. [Google Scholar] [CrossRef]
- Yayoshi-Yamamoto, S.; Taniuchi, I.; Watanabe, T. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol. Cell. Biol. 2000, 20, 6872–6881. [Google Scholar] [CrossRef] [Green Version]
- Grikscheit, K.; Frank, T.; Wang, Y.; Grosse, R. Junctional actin assembly is mediated by Formin-like 2 downstream of Rac1. J. Cell Biol. 2015, 209, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-L.; Liang, L.; Ding, Y.-Q. Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int. J. Colorectal Dis. 2008, 23, 1041–1047. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Zeng, Y.-F.; Guan, J.; Li, Y.-F.; Deng, Y.-J.; Bian, X.-W.; Ding, Y.-Q.; Liang, L. FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J. Pathol. 2011, 224, 377–388. [Google Scholar] [CrossRef]
- Chan, D.; Citro, A.; Cordy, J.M.; Shen, G.C.; Wolozin, B. Rac1 protein rescues neurite retraction caused by G2019S leucine-rich repeat kinase 2 (LRRK2). J. Biol. Chem. 2011, 286, 16140–16149. [Google Scholar] [CrossRef] [Green Version]
- Schreij, A.M.A.; Chaineau, M.; Ruan, W.; Lin, S.; Barker, P.A.; Fon, E.A.; McPherson, P.S. LRRK2 localizes to endosomes and interacts with clathrin-light chains to limit Rac1 activation. EMBO Rep. 2015, 16, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Nethe, M.; de Kreuk, B.J.; Tauriello, D.V.F.; Anthony, E.C.; Snoek, B.; Stumpel, T.; Salinas, P.C.; Maurice, M.M.; Geerts, D.; Deelder, A.M.; et al. Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts. J. Cell Sci. 2012, 125, 3430–3442. [Google Scholar] [CrossRef] [Green Version]
- Eide, P.W.; Cekaite, L.; Danielsen, S.A.; Eilertsen, I.A.; Kjenseth, A.; Fykerud, T.A.; Ågesen, T.H.; Bruun, J.; Rivedal, E.; Lothe, R.A.; et al. NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell. Signal. 2013, 25, 12–18. [Google Scholar] [CrossRef]
- Noda, Y.; Takeya, R.; Ohno, S.; Naito, S.; Ito, T.; Sumimoto, H. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 2001, 6, 107–119. [Google Scholar] [CrossRef]
- Cheung, L.W.T.; Yu, S.; Zhang, D.; Li, J.; Ng, P.K.S.; Panupinthu, N.; Mitra, S.; Ju, Z.; Yu, Q.; Liang, H.; et al. Naturally Occurring Neomorphic PIK3R1 Mutations Activate the MAPK Pathway, Dictating Therapeutic Response to MAPK Pathway Inhibitors. Cancer Cell 2014, 26, 479–494. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, F.; Li, X.; Song, X.; Zhou, L.; Jie, J. Screening key genes and miRNAs in early-stage colon adenocarcinoma by RNA-sequencing. Tumor Biol. 2017, 39, 1010428317714899. [Google Scholar] [CrossRef] [Green Version]
- Jezyk, M.R.; Snyder, J.T.; Gershberg, S.; Worthylake, D.K.; Harden, T.K.; Sondek, J. Crystal structure of Rac1 bound to its effector phospholipase C-beta2. Nat. Struct. Mol. Biol. 2006, 13, 1135–1140. [Google Scholar] [CrossRef]
- Klooster ten, J.P.; Leeuwen, I.V.; Scheres, N.; Anthony, E.C.; Hordijk, P.L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007, 26, 336–345. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Qiu, P.; Zhu, S.; Zhang, M.; Li, Y.; Zhang, M.; Wang, X.; Shang, J.; Qu, B.; Liu, J.; et al. SET nuclear proto-oncogene gene expression is associated with microsatellite instability in human colorectal cancer identified by co-expression analysis. Dig. Liver Dis. 2019. [Google Scholar] [CrossRef]
- Cristóbal, I.; Torrejón, B.; Rubio, J.; Santos, A.; Pedregal, M.; Caramés, C.; Zazo, S.; Luque, M.; Sanz-Alvarez, M.; Madoz-Gúrpide, J.; et al. Deregulation of SET is Associated with Tumor Progression and Predicts Adverse Outcome in Patients with Early-Stage Colorectal Cancer. J Clin Med 2019, 8, 346. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, S.C.; Schorr, A.; Roos, W.P.; Huelsenbeck, J.; Henninger, C.; Kaina, B.; Fritz, G. Rac1 protein signaling is required for DNA damage response stimulated by topoisomerase II poisons. J. Biol. Chem. 2012, 287, 38590–38599. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Xu, J.; Zhao, J.; Bai, J.H. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J. Cell. Biochem. 2018, 119, 7256–7263. [Google Scholar] [CrossRef]
- Kotelevets, L.; Trifault, B.; Chastree, E.; Scott, M.G.H. Posttranslational Regulation and Conformational Plasticity of PTEN. Cold Spring Harb. Perspect Med. 2020. [Google Scholar] [CrossRef]
- Diamantopoulou, Z.; White, G.; Fadlullah, M.Z.H.; Dreger, M.; Pickering, K.; Maltas, J.; Ashton, G.; MacLeod, R.; Baillie, G.S.; Kouskoff, V.; et al. TIAM1 Antagonizes TAZ/YAP Both in the Destruction Complex in the Cytoplasm and in the Nucleus to Inhibit Invasion of Intestinal Epithelial Cells. Cancer Cell 2017, 31, 621–634. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, Y.; Sagara, M.; Shibata, Y.; Shirouzu, M.; Yokoyama, S.; Akiyama, T. Identification and characterization of Asef2, a guanine-nucleotide exchange factor specific for Rac1 and Cdc42. Oncogene 2007, 26, 7620–7627. [Google Scholar] [CrossRef] [Green Version]
- Rosin-Arbesfeld, R.; Ihrke, G.; Bienz, M. Actin-dependent membrane association of the APC tumour suppressor in polarized mammalian epithelial cells. EMBO J. 2001, 20, 5929–5939. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.-Y.; Yu, Q.-F.; Shen, W.; Guo, C.-M.; Li, Z.; Zhou, X.-Y.; Zhou, N.-J.; Min, W.-P.; Gao, D. Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer. Int. J. Mol. Sci. 2016, 17, 2119. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-H.; Xia, D.; Kim, S.-W.; Holla, V.; Menter, D.G.; Dubois, R.N. Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells. Cancer Res. 2010, 70, 4054–4063. [Google Scholar] [CrossRef] [Green Version]
- Caspi, E.; Rosin-Arbesfeld, R. A novel functional screen in human cells identifies MOCA as a negative regulator of Wnt signaling. Mol. Biol. Cell 2008, 19, 4660–4674. [Google Scholar] [CrossRef] [Green Version]
- Gadea, G.; Blangy, A. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 2014, 93, 466–477. [Google Scholar] [CrossRef]
- Feng, J.; Zhao, J.; Xie, H.; Yin, Y.; Luo, G.; Zhang, J.; Feng, Y.; Li, Z. Involvement of NEDD9 in the invasion and migration of gastric cancer. Tumor Biol. 2015, 36, 3621–3628. [Google Scholar] [CrossRef]
- Abramovici, H.; Mojtabaie, P.; Parks, R.J.; Zhong, X.-P.; Koretzky, G.A.; Topham, M.K.; Gee, S.H. Diacylglycerol kinase zeta regulates actin cytoskeleton reorganization through dissociation of Rac1 from RhoGDI. Mol. Biol. Cell 2009, 20, 2049–2059. [Google Scholar] [CrossRef] [Green Version]
- Cai, K.; Mulatz, K.; Ard, R.; Nguyen, T.; Gee, S.H. Increased diacylglycerol kinase ζ expression in human metastatic colon cancer cells augments Rho GTPase activity and contributes to enhanced invasion. BMC Cancer 2014, 14, 208. [Google Scholar] [CrossRef] [Green Version]
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The biology of IQGAPproteins: Beyond the cytoskeleton. EMBO Rep. 2015, 16, 427–446. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Yang, Y.; He, Y.; Yang, P.; Wang, X.; He, G.; Zhang, P.; Zhu, H.; Xu, N.; Zhao, X.; et al. Cancer Letters. Cancer Lett. 2017, 411, 90–99. [Google Scholar] [CrossRef]
- Cheung, E.C.; Lee, P.; Ceteci, F.; Nixon, C.; Blyth, K.; Sansom, O.J.; Vousden, K.H. Opposing effects of TIGAR- and RAC1-derived ROS on Wnt-driven proliferation in the mouse intestine. Genes Dev. 2016, 30, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Carter, J.H.; Douglass, L.E.; Deddens, J.A.; Colligan, B.M.; Bhatt, T.R.; Pemberton, J.O.; Konicek, S.; Hom, J.; Marshall, M.; Graff, J.R. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin. Cancer Res. 2004, 10, 3448–3456. [Google Scholar] [CrossRef] [Green Version]
- Al-Maghrabi, J.; Emam, E.; Gomaa, W.; Al-Qaydy, D.; Al-Maghrabi, B.; Buhmeida, A.; Abuzenadah, A.; Al-Qahtani, M.; Al-Ahwal, M. Overexpression of PAK-1 is an independent predictor of disease recurrence in colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15895–15902. [Google Scholar]
- Gerçeker, E.; Boyacıoglu, S.O.; Kasap, E.; Baykan, A.; Yuceyar, H.; Yıldırım, H.; Ayhan, S.; Ellidokuz, E.; Korkmaz, M. Never in mitosis gene A-related kinase 6 and aurora kinase A: New gene biomarkers in the conversion from ulcerative colitis to colorectal cancer. Oncol. Rep. 2015, 34, 1905–1914. [Google Scholar] [CrossRef]
- Chow, H.Y.; Dong, B.; Valencia, C.A.; Zeng, C.T.; Koch, J.N.; Prudnikova, T.Y.; Chernoff, J. Group I Paks are essential for epithelial- mesenchymal transition in an Apc-driven model of colorectal cancer. Nat. Commun. 2018, 9, 3473. [Google Scholar] [CrossRef]
- Wang, X.; Gong, W.; Qing, H.; Geng, Y.; Wang, X.; Zhang, Y.; Peng, L.; Zhang, H.; Jiang, B. p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. Tumor Biol. 2010, 31, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.; Shulkes, A.; Baldwin, G.; He, H. Up-regulation of stem cell markers by P21- activated kinase 1 contributes to 5-fluorouracil resistance of colorectal cancer. Cancer Biol. Ther. 2016, 17, 813–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, G.; Henninger, C. Rho GTPases: Novel Players in the Regulation of the DNA Damage Response? Biomolecules 2015, 5, 2417–2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaya, K.; Oikawa, K.; Semba, S.; Tsuchiya, B.; Mukai, Y.; Otsubo, T.; Nagao, T.; Izumi, M.; Kuroda, M.; Domoto, H.; et al. Correlation between liver metastasis of the colocalization of actin-related protein 2 and 3 complex and WAVE2 in colorectal carcinoma. Cancer Sci. 2007, 98, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhou, M.; Xie, J.; Chao, P.; Feng, Q.; Wu, J. High glucose levels promote the proliferation of breast cancer cells through GTPases. Breast Cancer 2017, 9, 429–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, N.; Prieto, I.; Del Puerto-Nevado, L.; Portal-Nuñez, S.; Ardura, J.A.; Corton, M.; Fernández-Fernández, B.; Aguilera, O.; Gomez-Guerrero, C.; Mas, S.; et al. Diabetes Cancer Connect Consortium 2017 update on the relationship between diabetes and colorectal cancer: Epidemiology, potential molecular mechanisms and therapeutic implications. Oncotarget 2017, 8, 18456–18485. [Google Scholar] [CrossRef] [Green Version]
- Cannino, G.; Ciscato, F.; Masgras, I.; Sánchez-Martín, C.; Rasola, A. Metabolic Plasticity of Tumor Cell Mitochondria. Front. Oncol. 2018, 8, 333. [Google Scholar] [CrossRef]
- Kang, J.; Chong, S.J.F.; Ooi, V.Z.Q.; Vali, S.; Kumar, A.; Kapoor, S.; Abbasi, T.; Hirpara, J.L.; Loh, T.; Goh, B.-C.; et al. Overexpression of Bcl-2 induces STAT-3 activation via an increase in mitochondrial superoxide. Oncotarget 2015, 6, 34191–34205. [Google Scholar] [CrossRef]
- Han, J.; Theiss, A.L. Stat3: Friend or foe in colitis and colitis-associated cancer? Inflamm. Bowel Dis. 2014, 20, 2405–2411. [Google Scholar] [CrossRef] [Green Version]
- Valle-Mendiola, A.; Soto-Cruz, I. Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes. Cancers 2020, 12, 124. [Google Scholar] [CrossRef] [Green Version]
- Marei, H.; Malliri, A. Rac1 in human diseases: The therapeutic potential of targeting Rac1 signaling regulatory mechanisms. Small GTPases 2017, 8, 139–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado, M.D.M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Molecular Partners | Full Name | Function | Effect on Rac1 and Biological Impact | References Linked with CRC * |
|---|---|---|---|---|
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | small GTP-binding protein, Small GTPase | Rac1 oligomerization via C-terminal domain (basic amino acid residues 183–188). Increased intrinsic GTPase activity, increased PAK1 activation [11]. | [6,12] |
| RAC1b | Ras-related C3 botulinum toxin substrate 1, transcript variant Rac1b | small GTP-binding protein, Small GTPase | In frame insertion of an extra 19-amino acid sequence. Preferentially in a GTP-bound active form. Overexpressed in CRC. Not sufficient to initiate colorectal carcinogenesis in transgenic mice, but cooperates with Wnt signaling, and promotes colon carcinogenesis upon chronic inflammation. | [13,14,15] |
| Post Translational Modifications (PTMs); Phosphorylation | ||||
| AKT1, PKB | AKT Serine/Threonine Kinase 1 | Serine/Threonine Kinase | Rac1 inhibition: Akt phosphorylates Rac1 at Ser 71 and inhibits GTP-binding activity [16]. | [17,18] |
| MAPK3, ERK1, P44 | Mitogen-Activated Protein Kinase 3 | Member of the MAP kinase family, Serine/Threonine protein kinase | Phosphorylation of RAC1 at Thr108: decreased Rac1 activity, partially due to inhibiting its interaction with phospholipase C-γ1 (PLC-γ1) [19]. | [18,20,21] |
| PTK2, FAK1 | Protein Tyrosine Kinase 2 | Cytoplasmic protein Tyosine kinase concentrated at focal adhesions | Phosphorylation of Rac1 at Tyr64 decreases GTP binding and Rac1 activity. Phosphorylation targets Rac1 to focal adhesions. Decreased association with βPIX, and increased binding with RHOGDI. Decreased cell spreading [22]. | [23] |
| PRKCG | Protein Kinase C Gamma | Protein Serine/Threonine kinase activated by calcium and diacylglycerol | Phosphorylation of Rac1 at Ser71 promotes invasion of A431 cells [24]. | [25] |
| PRKCZ | Protein Kinase C Zeta | Member of the PKC family of Serine/Threonine kinase. Independent of calcium and diacylglycerol but not of phosphatidylserine | Phosphorylation of RAC1 at Ser71 promotes proliferation, and migration of LoVo colon cancer cells via PAK1/β-catenin pathway. | [26] |
| ROCK1 | Rho Associated Coiled-Coil Containing Protein Kinase 1 | Protein Serine/Threonine kinase, activated when bound to GTP-Rho | Phosphorylation of Rac1b at Ser71 facilitates its interaction with cytochrome c. Increased mitochondrial ROS production [27]. | [28] |
| SRC | SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase | Non receptor protein Tyrosine kinase | Phosphorylation of Rac1 at Tyr64 decreases GTP binding and Rac1 activity. Phosphorylation targets Rac1 to focal adhesions. Decreased association with βPIX, and increased binding with RhoGDI. Decreased cell spreading [22]. | [23,29] |
| SUMOylation/deSUMOylation | ||||
| PIAS3 | Protein Inhibitor of Activated STAT 3 | SUMO-E3 ligase | SUMOylation of Rac1 polybasic region (amino-acid residues 179–188) in the cytoplasm. Increased Rac1 activity and optimal cell migration in response to HGF signaling. PIAS3-mediated feedback loops controls cell proliferation and function as robust driving forces for Colitis Associated Cancer progression [30]. | [31] |
| TP53, P53 | Tumor Protein P53 | Tumor suppressor. Protein containing transcriptional activation, DNA binding, and oligomerization domains | Mutant TP53 competes with SENP1 for Rac1 interaction. Protects SUMOylated Rac1 from SENP1 peptidase. Increased Rac1 activity. In CRC, Rac1 SUMOylation/activation correlates with mutant TP53 status. | [32] |
| SENP1 | SUMO Specific Peptidase 1 | Cysteine protease, deconjugates sumoylated proteins. | Mutant TP53 competes with SENP1 for Rac1 interaction. Decreases Rac1 SUMOylation and activity [32]. | |
| Ubiquitination | ||||
| HACE1 | HECT Domain and Ankyrin Repeat Containing E3 Ubiquitin Protein Ligase 1 | E3 ubiquitin-protein ligase. Tumor suppressor. Mutated in cancers | Ubiquitinates and targets Rac1 (preferentially GTP-bound form) to proteasomal degradation. The Rac1 downstream kinase PAK phosphorylates and impairs HACE1 -induced Rac1 ubiquitination. Controls cell proliferation, cell migration, ROS production [33,34,35,36,37]. | [38,39] |
| FBXL19 | F-Box and Leucine Rich Repeat Protein 19 | E3 ubiquitin ligases (member of the Skp1-Cullin-F-box family) | Ubiquitination of Lys 166 of Rac1 (preferentially phosphorylated at Ser71). Rac1 degradation by proteasome [40]. | |
| BIRC2, c-IAP1 | Baculoviral IAP Repeat Containing 2 | E3 ubiquitin-protein ligase. Member of family of apoptotic suppressor proteins | Polyubiquitination of activated Rac1 at Lys147 and proteasomal degradation. Reverses mesenchymal morphology and cell migration [41,42]. | [43] |
| XIAP | X-Linked Inhibitor of Apoptosis | E3 ubiquitin-protein ligase. Member of family of apoptotic suppressor proteins | Polyubiquitination of activated Rac1 at Lys147 and proteasomal degradation. Reverses mesenchymal morphology and cell migration [41,42]. | [43] |
| Bacterial Toxins-Induced PTMs | ||||
| Toxin | Source | Rac1 PTM | Effect on Rac1 and Biological Impact | |
| TcdA, TcdB (toxin A, B) | Clostridium difficile | Mono-glycosylation of Thr35 of Rac1 (switch I). | Rac1 inactivation, actin depolymerisation, loss of cell-cell contacts and apoptosis [44] | |
| VopS | Vibrio parahaemolyticus | AMPylation of Thr35 of Rac1 (switch I). | Rac1 inactivation [45]. | |
| YopT | Yersinia enterocolitica | Cysteine protease; proteolysis of the Rac1 carboxy-terminal upstream CAAX box | Rac1 inactivation. Impaired membrane interaction [46]. | |
| VopC; DNT (dermonecrotic toxin); CNF1 | Vibrio parahaemolyticus; Bordetella dermonecrotizing; Escherichia coli | Deamidation Gln61 of Rac1 (switch II) Gln -> Glu | Constitutive Rac1 activation (decreased Rac1 GTPase activity) [47]. | |
| DNT (dermonecrotic toxin) | Bordetella dermonecrotizing | Polyamination of Gln61 (evidenced in vitro, not confirmed in vivo) | Constitutive Rac1 activation (decreased Rac1 GTPase activity). Note that DNT also deaminates Gln61 of Rac1 [48] | |
| ExoS (Exoenzyme S) | Pseudomonas aeruginosa | ADP-ribosylation of Arg 66 and Arg 68; cell line dependent (HT29 colon cancer cells sensitive; J774A.1 macrophages unsensitive) | Rac1 activation. Note that exoS ADP-ribosyltransferase activity is C-term, an antagonistic GTPase-activating domain is in the N-term of ExoS [49]. | |
| Molecular Partners | Full Name | Function | Effect on Rac1 and Biological Impact | References Linked with CRC * |
|---|---|---|---|---|
| RAC1 Regulators: RhoGAPs, RhoGDIs, GEFs, RhoGAPs, RhoGDIs | ||||
| ARHGAP1 | Rho GTPase-activating protein 1 (Rho-related small GTPase protein activator) (p50-RhoGAP) | GTPase activating protein. Negative regulation of Rho GTPases | Acting as a negative regulator for Rac1-mediated signaling [123]. | |
| ARHGAP15 | Rho GTPase Activating Protein 15 | GTPase activating protein. Negative regulation of Rho GTPases | Decreases RAC1 activity. ARHGAP15 inhibits growth, migration, and invasive properties of the human colonic cell lines. Downregulated in CRC. | [124] |
| ARHGAP22 | Rho GTPase Activating Protein 22 | GTPase activating protein. Negative regulation of Rac1 and Cdc42 GTPases | Rho signaling via ROCK to ARHGAP22 inactivates Rac1, suppresses mesenchymal movement and promotes amoeboid movement [9]. | |
| ARHGAP24, FILGAP | Rho GTPase Activating Protein 24 | Rac1 specific GTPase activating protein | Filamin A targets ARHGAP24 to membrane protrusion, where it antagonizes Rac1. This leads to suppression of lamellae formation and promotion of retraction. Downregulated in CRC [125]. | [126] |
| ARHGAP32 | Rho GTPase Activating Protein 32; (Brain-specific Rho GTPase-activating protein) (RICS) | GTPase-activating protein (GAP) promoting GTP hydrolysis on RhoA, Cdc42 and Rac1 small GTPases; constitutively associated with TrkA, a high-affinity receptor for NGF | Stimulates GTP hydrolysis of RhoA and Cdc42 over that of Rac1. Components in NGF-induced cytoskeletal; regulates neurite outgrowth by modulating Rho family of small GTPases; possible role in in neural/glial cell proliferation; activity regulated by Src-like tyrosine kinases [127,128,129]. | [130,131] |
| ARHGAP43/SH3BP1 | SH3 Domain Binding Protein 1 | GTPase activating protein. Negative regulation of Rac1 and Cdc42 GTPases. Partner of the exocyst complex (tethers secretory vesicles to the plasma membrane) | Stimulates the GTPase activity of Rac1 during migration. Localizes together with the exocyst to the leading edge of motile cells. Rac1 inactivation is required to organize protrusions and to drive efficient directional motility. SH3BP1 overexpression promoted invasion, migration, and chemoresistance of cervical cancer cells through increasing Rac1 activity and Wave2 protein level. This effect might be related to cross-talk between Ral and Rac1 pathways [132,133,134]. | |
| RACGAP1 | Rac GTPase Activating Protein 1 | Negative regulation of Rho GTPases | Decreases RAC1 activity. Overexpressed in colorectal cancers. Nuclear localization associated with poor patient outcome. | [135,136] |
| ARHGDIA, Rho GDI 1 | Rho GDP Dissociation Inhibitor Alpha, Rho GDP-dissociation inhibitor 1 | Regulates GDP/GTP exchange of Rho GTPases: inhibits dissociation of GDP. | Regulates the GDP/GTP exchange cycle and sequesters Rho GTPases in the cytosol as an inactive pool. [137,138,139]. | [140,141] |
| GDP/GTP Exchanges, Guanine Nucleotide Exchanges | ||||
| ARHGEF6/COOL-2/alpha-Pix | Rac/Cdc42 guanine nucleotide exchange factor 6 (alfa-Pix) (COOL 2) (Pak-interactive exchange factor) | Rac-specific guanine nucleotide exchange factor (GEF) | Dimerization enables its Dbl and pleckstrin homology domains to work together to bind specifically to Rac-GDP. Dimer dissociation into monomeric form allows it to act as a GEF for Cdc42 and Rac1. ARHGEF6 was identified as one of key genes and pathways in the pathogenesis of CRC (microarray data analysis) [142]. | [143] |
| ARHGEF7/ COOL-1/ beta-PIX | Rho guanine nucleotide exchange factor 7 (Beta-Pix) (COOL-1) (PAK-interacting exchange factor beta) (p85) | Rac1-specific guanine nucleotide exchange factor (GEF) | C-terminal portion of the PH domain of COOL-1 works together with the DH domain through dimerization to mediate highly specific interaction with GDP-bound Rac1 via SH3 domain of β-Pix. Recruits Rac1 to membrane ruffles and to focal adhesions. Rac1 activation mediates cell spreading. Increased expression of ARHGEF7 in CRC associated with metastasis. ARHGEF7 mutations associate with worse disease-free survival. ARHGEF 7 is one of determinants for irinotecan sensitivity/resistance in colorectal cancer cell lines [144] | [145,146,147] |
| ARHGEF33 | Rho guanine nucleotide exchange factor 33 | Rho guanine nucleotide exchange factor (GEF) | ARHGEF33 may play a similar role to KRAS and NRAS mutations. | [146] |
| ARHGEF4/Asef1 | Rho Guanine Nucleotide Exchange Factor 4 | Guanine nucleotide exchange factor for RhoA, Rac1 and cdc42. Binding of APC activates Rac1 GEF activity | Rac1 activation. Lamellipodia formation, and increased cell migration. Mutated APC and Asef are involved in the migration of colorectal tumor cells. | [148,149] |
| ARHGEF29/SPATA13/Asef2 | Rho Guanine Nucleotide Exchange Factor 29 | Guanine nucleotide exchange factor for RhoA, Rac1 and cdc42. Binding of APC activates RAC1 GEF activity | RAC1 activation. Asef2 is required for migration of colorectal tumor cells expressing truncated APC. Asef1 and Asef2 are required for adenoma formation in Apc(Min/+)mice. The aberrant subcellular localization of these complexes in CRC cells may contribute to the cells aberrant adhesive and migratory properties. | [148,149,150] |
| DOCK1, DOCK180 | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180) | Guanine nucleotide exchange factor (GEF) The complex formation between Dock180 and Elmo1 acts as a bipartite guanine nucleotide exchange factor for Rac1. | Activator of Rac1. CrkII/Dock180/Rac1 pathway involved in integrin signaling. ELMO-DOCK180 complex is positioned at the membrane to activate Rac1 signaling and actin cytoskeleton remodeling, phagocytosis and motility. Dock1 plays critical roles in receptor tyrosine kinase -stimulated cancer growth and invasion. Dock180 activity is required in cell migration and tumorigenesis promoted by PDGFR and EGFR. The cortactin-driven (CTTN) invasion by CRC cells is dependent of the activation of DOCK1-Rac1 [151,152,153,154,155,156,157,158,159]. | [160,161] |
| DOCK2 | Dedicator of cytokinesis protein 2 | Guanine nucleotide exchange factor. Predominantly expressed in peripheral blood leukocytes | Regulators of Rac1 function in adherent and non-adherent cells. Dock/ELMO complex functions as an unconventional two-part exchange factor for Rac1. ELMO binding to the SH3 domain of Dock2 disrupted the SH3: Docker interaction, facilitating Rac1 access to the Docker domain, and contributes to the GEF activity of the Dock2/ELMO complex. High mutation prevalence of DOCK2 in CRC at frequencies of >7%. More frequently mutated in hypermutated CRC. High expression of DOCK2 related with good prognosis [159,162]. | [130,163,164] |
| DOCK3/MOCA | Dedicator of Cytokinesis 3, modifier of cell adhesion protein | Rac1 GEF. Rac1-binding domain assigned to amino acids 939 to 1323. C-term region required for optimal binding | DOCK3 activates Rac1 leading to cytoskeletal reorganization. NEDD9 complexes with DOCK3 to regulate Rac1 activity. A genome-wide significant association of rs17659990 intronic variant with CRC risk [9,165]. | [166] |
| DOCK4 | Dedicator of Cytokinesis 4 | Rac1 GEF. Induces GTP loading of Rac1 | DOCK4 acts as a scaffold protein in the Wnt/β-catenin pathway. DOCK4 is required for Wnt-induced Rac1 activation, TCF transcription and cell migration. DOCK4 overexpression is associated with resistance of CRC cell lines to Cetuximab and Trastuzumab [159]. | [167,168,169] |
| DOCK7 | Dedicator of Cytokinesis Protein 7 | Rac1 and Rac3 specific GEF. | Activator of Rac GTPases. Regulation of microtubule networks, axon formation and neuronal polarization [170,171]. | |
| ELMO1 | Engulfment and Cell Motility 1 | ELMO is a critical regulator of DOCKs1-5. Dock-ELMO complex functions as a bipartite GEF for Rac1. | ELMO binding to the SH3 domain of DOCKs allows Rac1 access to the Docker domain, and contributed to the GEF activity of the DOCK/ELMO complex [158,159,172]. | |
| FARP2 | FERM, ARH/RhoGEF And Pleckstrin Domain Protein 2 | Rac1 guanine nucleotide exchange factor (GEF) | RAC1 activation. Collective invasion of colorectal cancer cells. | [173] |
| KALRN | Kalirin RhoGEF Kinase | Rho guanine nucleotide exchange factors (GEF) | GDP/GTP exchange, Rac1 activation. Induces neurite initiation, axonal growth, and dendritic morphogenesis. Promotes smooth muscle cells migration and proliferation. Paralog of TRIO [174,175,176]. | |
| NEDD9 | Neural precursor cell-expressed, developmentally-downregulated 9, also called HEF1 and Cas-L | Adaptor molecule, member of the p130Cas family | Skeleton protein, modulates invasion, metastasis, proliferation and apoptosis of tumor cells. NEDD9 complexes with DOCK3 to regulate Rac1 activity. Blocking Rac1 signaling suppresses mesenchymal migration and enhances amoeboid movement. Downregulation of NEDD9 by apigenin suppresses migration, invasion, and metastasis of CRC cells. Elevated expression in CRC correlates with high TNM stage and associated with poor prognosis [9,177,178]. | [130,163,179,180] |
| PREX1 | Phosphatidylinositol-3,4,5-Trisphosphate Dependent Rac Exchange Factor 1 | Guanine-nucleotide exchange factors for Rac family small G proteins | Stimulation of cell migration [181,182]. | |
| PLCG1, PLC-gamma-1 | Phospholipase C Gamma 1 | Hydrolysis of phosphatidylinositol 4,5-bisphosphate to 1,4,5-trisphosphate (IP3) and diacylglycerol | Critical for EGF-induced Rac1 activation in vivo. PLC-g SH3 domain acts as a Rac1 guanine nucleotide exchange in vivo. Cytoskeleton remodeling and cell migration [183]. | |
| RCC2 | Regulator of Chromosome Condensation 2 | Guanine exchange factor for RalA | Interacts with Switch regions. RCC2 inhibits GEF-mediated activation of Rac1, preventing formation of multiple protrusions. RCC2 interacts with and transfers Rac1-GDP to Coro1C. Depletion of Coro1C or RCC2 causes loss of cell polarity that results in shunting migration. RCC2 is a transcriptional target of TP53, it deactivates Rac1 and inhibits migration of colon cancer cells. Decreased expression in CRC associated with a good prognosis for patients with MSI-CRC, a poor prognosis for patients with MSS-CRC [184]. | [185,186] |
| SOS1 | Son of sevenless homolog 1. Ras/Rac Guanine Nucleotide Exchange Factor 1 | Guanine nucleotide exchange factor (GEF) for Ras and Rac1 | Sos1 triggers Rac1 GDP/GTP exchange and activates Rac1 and JNK, leading to membrane ruffling [187]. | |
| TIAM1 | T Cell Lymphoma Invasion and Metastasis | Rac1-specific guanine nucleotide exchange factor (GEF) | Rac1 activation. Stabilization of junctional complexes. Actin cytoskeleton remodeling, membrane ruffling, cell motility, invasiveness. Neurite outgrowth. Overexpressed in CRC, associated with poor prognosis and resistance to chemotherapies [188,189,190]. | [53,191,192,193,194,195,196,197,198] |
| VAV1 | Vav Guanine Nucleotide Exchange Factor 1 | Guanine nucleotide exchange factors (GEFs). Expressed in hematopoietic cells | Overexpression is associated with human CRC at advanced stage and with lymph node metastasis [199]. | [200] |
| VAV2 | Vav Guanine Nucleotide Exchange Factor 2 | Guanine nucleotide exchange factors (GEFs). Ubiquitous | Mutations associated with high risk of recurrence for patients with stages II and III CRC [199]. | [201] |
| Molecular Partners | Full Name | Function | Effect on Rac1 and Biological Impact | References Linked with CRC * |
|---|---|---|---|---|
| RAC1 Downstream Effectors; Nitric Oxide (NO), Reactive Oxygen Species (ROS) Production | ||||
| NOXA1 | NADPH oxidase activator 1, P67phox-Like Factor | Cytosolic subunit recruited to the plasma membrane to form the active NADPH oxidase complex (NOX1) with p22phox, NOXO1, Rac1 and NADPH oxidase1 that produces the superoxide anion | Activated Rac-1 interacts with NOXA1 in the NOX1 holoenzyme at the membrane leading to reactive oxygen species production involved in cell signaling and innate immune response [202]. | |
| NCF2, P67-Phox | Neutrophil cytosol factor 2 (NCF-2) (67 kDa neutrophil oxidase factor) (NADPH oxidase activator 2) (Neutrophil NADPH oxidase factor 2) (p67-phox) | Rac-GTP binds to the N-terminal tetratricopeptide repeat domain of p67phox. Cytosolic subunit is recruited to the membrane to form the active NADPH oxidase complex (NOX2) with p22phox, p40phox, p47phox, Rac1 (monocytes, Rac2 neutrophils) and gp91phox that produces the superoxide anion | Regulation of phagocytic NADPH oxidase activity. Activated Rac-1 interacts with p67-Phox in the NADPH complex at the membrane, causes a conformational change in the "activation domain" in p67-Phox leading to reactive oxygen species production and innate immune response [203,204,205,206,207,208]. | |
| APEX1/APE1 | Apurinic/Apyrimidinic Endodeoxyribonuclease 1 | Endonuclease. Involved in DNA repair and redox regulation of transcriptional factors | Interaction with RAC1 impairs NOX1 complex formation. Decreased ROS production [81]. | |
| BCL2 | BCL2 Apoptosis Regulator | Integral outer mitochondrial membrane protein. Inhibitor of apoptotic death | Increased ROS production. Mitochondrial oxidative stress. Although anti-apoptotic molecule, a meta-analysis suggests that Bcl-2 is for a favorable prognosis for patient with CRC [209]. | [210] |
| CYCS | Cytochrome c | Central component of the electron transport chain in inner membrane of mitochondria | Mitochondrial ROS production. Electron transfer from cytochrome c to Rac-1 modulates mitochondrial H2O2 production. Rac1b phosphorylation at Ser71 facilitates its interaction with cytochrome c. Rac1B may be involved in Hutchinson-Gilford progeria syndrome. [27,211]. | |
| NOS2 | Nitric oxide synthase-2 | Inducible by LPS and certain cytokines, generates nitric oxide mediator | Increased nitrite generation and NOS2 activity through subcellular redistribution [212]. | |
| Transcription (co)Factors | ||||
| CTNNB1 | Catenin Beta 1, Beta-catenin | Component of the complex of proteins that constitute adherens junctions: interacts with E-cadherins. Component of the canonical Wnt signaling pathway: In the presence of Wnt ligand or consecutively to mutations of components of the complexes that trigger its phosphorylation and degradation by the proteasome (e.g., APC), β-catenin translocates in the nucleus, and acts as a coactivator for transcription factors of the TCF/LEF family, leading to transcription of Wnt target genes (e.g., Myc, Cyclin D1, MMP7) | Rac-1 interacts with β-catenin through its polybasic region. In the absence of APC in mouse intestinal epithelium, Rac1 is not required for β-catenin nuclear localization and/or for its functional activity. In LoVo colon cancer cells (mutant APC), Rac1 phosphorylation at Ser71 by PKCZ promotes nuclear β-catenin accumulation through PAK1 activation. In NIH3T3, SW480 and HCT116 cells, Rac-1 silencing or overexpression do not influence this nuclear accumulation. In contrast upon Wnt stimulation, active Rac1 induces the redistribution of Rac1/ β-catenin protein complex from the plasma membrane to the nucleus, favors the formation of β-catenin/LEF1 complex and potentiates the transactivation of Wnt responsive genes, via Jnk -induced β-catenin phosphorylation. These discrepancies might originate from the level of Rac1 accumulation and activity, and from the cellular context [26,56,58,213,214] | [3,4] |
| DVL3 | Dishevelled Segment Polarity Protein 3 | Cytoplasmic phosphoprotein involved in Wnt signaling pathway | RAC1b interacts with Dishevelled-3 to form a tetramer with β-catenin/TCF. Transcription of canonical Wnt target genes [112]. | |
| STAT3 | Signal Transducer and Activator of Transcription 3 | Transcription factor activated by receptor associated kinases | Activated Rac1 stimulated STAT3 phosphorylation on both tyrosine and serine residues. Epithelial-mesenchymal transition and invasion of CRC cells. Overexpressed in CRC [215]. | [89,216] |
| UNKL, Unkempt | Unk Like Zinc Finger | Contributes to E3 ligase activity. Nuclear localization | Activated Rac1 interacts with UNKL and promotes ubiquitination of BAF60b, a component of SWI/SNF chromatin remodeling complexes involved in regulation of transcription and chromatin remodeling [217]. | |
| Kinases | ||||
| PAK1 | P21 (RAC1) Activated Kinase 1 | Serine/threonine-protein kinase PAK1, belongs to the subgroup I of PAKs. Interacts with the p21-binding domains PBDs of Rac1 | PAK1 is activated upon binding to GTP-Rac1. Implicated in cytoskeleton dynamics, cell adhesion, migration, proliferation, apoptosis, mitosis, DNA Damage Repair, and vesicle-mediated transport processes. PAK1 phosphorylates Bad leading to uncoupling of Bad/Bcl-2 and enhanced cell survival; effectors of cytoskeletal reorganization. Links Rac1 to JNK/MAPK pathways: phosphorylates and activates MAP2K1, RAF1. Regulates transcription through association and/or phosphorylation of transcription factors, co-regulators and cell cycle-related proteins. PAK1 phosphorylation of Snail favors its nuclear accumulation and promotes transcriptional repression of E-cadherin. Phosphorylation of NF-kB triggers nuclear translocation and the transcriptional activity of the p65 subunit. Implicated in mediating signaling from Rac1 to JNK and to actin cytoskeleton. PAK1 expression is associated with CRC metastasis [123,218,219,220,221,222]. | [219,220] |
| PAK4 | p21 protein (Cdc42/Rac)-activated kinase 4 Serine/threonine-protein kinase PAK4 | Serine/threonine-protein kinase PAK4, Belongs to the subgroup II of PAKs. Interacts with the p21-binding domains PBDs of Rac1 | PAK4 nuclear accumulation enhances β-catenin nuclear import and increases TCF/LEF transcriptional activity. PAK4 expression is associated with CRC metastasis. | [219,220,223] |
| PAK5 | p21 protein (Cdc42/Rac)-activated kinase 5 Serine/threonine-protein kinase PAK5 | Serine/threonine-protein kinase PAK5, belongs to the subgroup II of PAKs, mitochondrial localization | Overexpressed in CRC, correlates with tumor stage and dedifferentiation. | [224] |
| MAP3K1, MEKK1 | Mitogen-Activated Protein Kinase Kinase Kinase 1 | Serine/threonine kinase. Involved in signal transduction cascades of ERK and JNK kinase and NF-kappa-B pathways. Nuclear and post-Golgi vesicle-like compartment | Interacts with active GTP-bound Rac1. Activation of the Erk and JNK kinases via MAP2K1 and MAP2K4 [225]. | |
| MAP3K10, MLK2 | Mitogen-Activated Protein Kinase Kinase Kinase 10 | Serine/threonine kinase. Activates MAPK8/JNK and MKK4 pathway | CRIB domain interacts with GTP-bound form of Rac1. Activates MAPK8/JNK and MKK4/SEK1 [226]. | |
| MAP3K4, MEKK4 | Mitogen-Activated Protein Kinase Kinase Kinase 4 | Serine threonine kinase. Activates MAPK14 (P38alpha) and JNK pathways, but not ERK | Interacts with active GTP-bound Rac1. JNK activation [225,227]. | |
| MAP3K11, MLK3 | Mitogen-Activated Protein Kinase Kinase Kinase 11 | Activates MAPK8/JNK and NF-kappaB transcriptional activity | CRIB domain interacts with GTP-bound form of Rac1. JNK activation [226]. | |
| Scaffolding Molecules/ Rac1 Subcellular Targeting/Cytoskeletton Remodeling, Membrane Ruffling | ||||
| IQGAP1, P195 | IQ Motif Containing GTPase Activating Protein 1; Ras GTPase-activating-like protein IQGAP1 | Scaffolding molecule. Regulates dynamics and assembly of actin cytoskeleton | Activated Rac1/Cdc42, IQGAP1, and CLIP-170 form a tripartite complex; activated Rac1 recruits MTs through IQGAP1. Role in cell polarization, actin crosslinking protein, accumulates at the polarized leading edge and areas of membrane ruffling. Overexpressed in tumor tissues as compared with control mucosa; localized at invasion front [228,229,230]. | [231] |
| IQGAP2 | Ras GTPase-activating-like protein IQGAP2 | Scaffolding molecule. Dynamics and assembly of actin cytoskeleton | Role in generation of specific actin structures; subcellular Rac1 localization. Expression in CRC correlates positively with patient survival [232]. | [233] |
| IQGAP3 | Ras GTPase-activating-like protein IQGAP3 | Scaffolding molecule. Dynamics and assembly of actin cytoskeleton | Overexpressed in MSS TP53 mutant CRC, expression levels correlated inversely with survival. | [233,234] |
| SH3RF1, POSH | SH3 Domain Containing Ring Finger 1, Plenty of SH3 Domains | Scaffolding molecules for components of the JNK signaling pathway. E3 ubiquitin-protein ligase activity | Links activated Rac1 and downstream JNK kinase cascade (MLKs, MLK4/7, JNK1/2). Induction apoptotic cascade [235,236]. | |
| SH3RF3, POSH2 | SH3 Domain Containing Ring Finger 3 | Scaffolding molecules for components of the JNK signaling pathway. E3 ubiquitin-protein ligase activity | Scaffold for a multiprotein complex that transduces signals from GTP-loaded Rac1 to JNK activation [237]. | |
| CCM2 | CCM2 Scaffold Protein, osmosensing scaffold for MEKK3 | Involved in stress-activated p38 Mitogen-activated protein kinase signaling cascade | Binds to actin, Rac1, MEKK3 and MKK3. Role in osmoregulation [238]. | |
| KPNA2 | Karyopherin alpha2, importin α-1 | Nuclear transport of proteins (binds NLS) | Interacts with Rac1 nuclear localization signal (NLS, C-terminal polybasic region). Nuclear import, (independent GDP/GTP loading), requires Rac1 activation. Nuclear Rac1 coimmunoprecipitates with numerous proteins. Overexpressed in CRC [239]. | [240] |
| YWHA, 14-3-3 | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein | Binds to phosphoserine-containing proteins | Rac1 Ser71 phosphorylation increases affinity for 14-3-3 proteins. Interaction increases EGF -induced Rac1 activation. Cytoplasmic localisation of the complexe. Overexpressed in CRC [93]. | [241] |
| RAP1GDS1, SmgGDS | Rap1 GTPase-GDP dissociation stimulator 1 (Exchange factor smgGDS) (SMG GDS protein) (SMG P21 stimulatory GDP/GTP exchange protein) | C-terminal poly-basic region of Rac1 | SmgGDS is a GEF for RhoA and RhoC, but not for Rac1. SmgGDS interacts with RAC1 C-terminal polybasic region and triggers Rac1 nuclear translocation and degradation by proteasome. Rac1b interacts more efficiently to SmgGDS than Rac1. SmgGDS splice variants control Rac1 prenylation and membrane localization: SmgGDS-558 associates with prenylated Rac1, SmgGDS-607 associates with nonprenylated GTPases and regulates its entry into the prenylation pathway [111,242,243,244]. | |
| ARFIP2/POR1 | Arfaptin-2 (ADP-ribosylation factor-interacting protein 2) (Partner of RAC1) (Protein POR1) | Downstream effector, plasma membrane | POR1 binds preferentially to GTP-bound form of Rac1 (effector binding domain, aminoacid residues 26-48). Mediates Rac1-induced signals; membrane ruffling; regulates the organization of the actin cytoskeleton [245,246,247]. | |
| BAIAP2, IRSp53 | BAI1 Associated Protein 2 | Adaptor protein; links membrane bound G-proteins to cytoplasmic effector proteins | RAC1-mediated membrane ruffling. Involved in the regulation of the actin cytoskeleton by WASF family members and the Arp2/3 complex [248]. | |
| CTNND1/P120ctn | Catenin Delta 1/P120 catenin | Member of the Armadillo protein family. Involved in cell adhesion (binds E-cadherin) and transcription | P120 catenin interacts with the extra 19-amino acid sequence of RAC1b. Directed cell movement [111]. | |
| CAV1 | Caveolin-1 | Scaffolding protein, caveolar membranes | Coronin-1C and caveolin retrieve Rac1 from similar locations at the rear and sides of the cell. In absence of fibronectin, Coronin-1C-mediated Rac1-GDP extraction and recycling to the leading edge, and maintains Rac1 cellular levels. In absence of coronin-1C, caveolin-mediated endocytosis targets Rac1 for proteasomal degradation, consecutively to engagement of the fibronectin receptor syndecan-4 [249]. | |
| CORO1C | Coronin 1C | Member of the WD repeat protein family. Actin-binding proteins that regulate actin branching by inhibition of the Arp2/3 complex and stimulation of actin depolymerization by cofilin | Coro1C Binds Rac1-GDP. Release inactive Rac1 from non-protrusive membrane. Required for Rac1 redistribution to a protrusive tip and fibronectin-dependent Rac1 activation. Increases accumulation of activated RAC1 at the leading edge of migrating cells. Directional fibroblast migration [184,249]. | |
| CYFIP1, SRA-1 | Cytoplasmic FMR1 Interacting Protein 1 | Regulates cytoskeletal dynamics and protein translation. Component of the WAVE regulatory complex (WRC), through actin polymerization | RAC1 binds to CYFIP1, initiating WASF3 complex formation involved in cytoskeleton reorganization and polymerization. Promotes invasiveness of breast prostate and colon cancer cell lines [250,251]. | |
| FLNA | Filamin A | Actin binding protein, connects cell membrane constituents to actin cytoskeleton | Mechanical force through β1-integrins triggers apoptosis through Rac1/Pak1/p38 signaling pathway. FLNa recruits ARHGAP24 to sites of force application suppressing Rac1 activation, lamallae formation and Rac1/p38-mediated apoptosis. Down-regulated in CRC. | [252,253] |
| FLNB | Filamin B | Actin binding protein, connects cell membrane constituents to actin cytoskeleton | JNK activation and induction of apoptosis in response to type I Interferon [254,255]. | |
| FMNL1, FRL | Formin Like 1 | Actin polymerization, morphogenesis, cytokinesis, and cell polarity | Interacts preferentially with GTP-bound Rac1. Regulation of motility and survival of macrophages [256]. | |
| FMNL2 | Formin Like 2 | Actin polymerization, morphogenesis, cytokinesis, and cell polarity | Activation of Rac1 steers FMNL2 to de novo junctional actin formation at newly formed cell-cell contacts and adherens junction formation. FMNL2 enhances proliferation, motility and invasiveness of colon cancer cell lines. Overexpressed in CRC and in liver metastases [257]. | [258,259] |
| LRRK2 | Leucine Rich Repeat Kinase 2 | Member leucine-rich repeat kinase family (protein with ankryin repeat region, leucine-rich repeat domain, kinase domain, DFG-like motif, RAS domain, GTPase domain, MLK-like domain, and WD40 domain). Mutation causes dominant-inherited Parkinson’s disease | Overexpression and knockdown of LKRR2 simulates Rac1 activity. Role in maintenance of neurite morphology. Role in synaptic vesicle trafficking [260,261]. | |
| NEDD4-1 | Neural Precursor Cell Expressed, Developmentally Down-Regulated 4, E3 Ubiquitin Protein Ligase | E3 Ubiquitin Protein Ligase | Rac1 stimulates Nedd4 activity and increases ubiquitylation and degradation of the adapter protein dishevelled-1 that transduces Wnt signal downstream frizzled receptor. Maturation of epithelial cell-cell contacts. Overexpressed in CRC. [262]. | [263] |
| PARD6A, PARD6B, PARD6G | Par-6 Family Cell Polarity Regulator Alpha/Beta/Gamma | Protein with a PDZ domain and a semi-Cdc42/Rac interactive binding (CRIB) domain. Involved in asymmetrical cell division and cell polarization | Interacts with GTP-bound Rac1. PAR6, Rac1 and atypical PKC colocalize as a ternary complex in membrane ruffles (leading edge of polarized cells during movement) [264]. | |
| PIK3R1, P85A | Phosphoinositide-3-Kinase Regulatory Subunit 1 | Regulatory subunit of PI3K | P85 binds GTP bound Rac1. Rac1 triggers P85a nuclear translocation. Activation of ERK and JNK Signaling Cascades. Downregulated in CRC [265]. | [4,17,266] |
| PLCB2 | Phospholipase C Beta 2 | Hydrolysis of phosphatidylinositol 4,5-bisphosphate to 1,4,5-trisphosphate (IP3) and diacylglycerol | Rac1 engages the PH domain of PLC-beta2 and optimizes its orientation for substrate membranes. Increased PLC activity [267]. | |
| SET | SET nuclear proto-oncogene; protein phosphatase type 2A inhibitor | Inhibits acetylation of nucleosomes (especially histone H4) | SET potentiates Rac1-mediated cell migration. Phosphorylation at Ser9 dissociates SET dimers and allows SET redistribution from nucleus to cytoplasm. The SET/activated Rac1 complex is recruited to the plasma membrane, and stimulates kinase-mediated signaling. SET enhances cell migration, EMT, and induces MYC expression in CRC cells. Overexpressed is in early-stage CRC, associated with progression and aggressiveness, and a poor outcome. Lower levels of SET in MSI CRCs compared to MSS CRC [268]. | [269,270] |
| TOP2A | DNA Topoisomerase II Alpha | Catalyzes transient breaking and rejoining of two strands of duplex DNA to relieve torsional stress that occurs during DNA transcription and replication | Rac1 is required for DNA damage induction and subsequent activation of DNA Damage Repair following treatment with topo II inhibitors. Overexpressed in CRC. Knowdown in CRC cells decreases Akt and Erk activity, and suppresses cell proliferation and invasion [269,271]. | [272] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotelevets, L.; Chastre, E. Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis. Cancers 2020, 12, 665. https://doi.org/10.3390/cancers12030665
Kotelevets L, Chastre E. Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis. Cancers. 2020; 12(3):665. https://doi.org/10.3390/cancers12030665
Chicago/Turabian StyleKotelevets, Larissa, and Eric Chastre. 2020. "Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis" Cancers 12, no. 3: 665. https://doi.org/10.3390/cancers12030665
APA StyleKotelevets, L., & Chastre, E. (2020). Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis. Cancers, 12(3), 665. https://doi.org/10.3390/cancers12030665