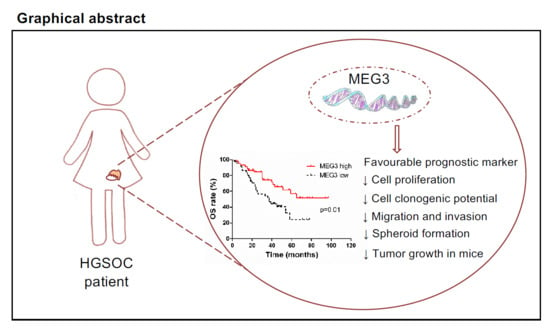
Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer
Abstract
:1. Introduction
2. Results
2.1. MEG3 Was Found to Be an Independent Favorable Prognostic Factor in Advanced HGSOC
2.2. MEG3 Expression in HGSOC Cell Lines
2.3. MEG3 Regulated the Proliferation of HGSOC Cells
2.4. MEG3 Overexpression Inhibited Cell Migration and Invasion of HGSOC Cells
2.5. MEG3 Overexpression Inhibited Spheroid Growth in Extracellular Matrix
2.6. MEG3 Overexpression Inhibited the Growth of HGSOC in Mice
2.7. MEG3 Overexpression Upregulated PTEN
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients
4.2. RNA Extraction from FFPE Tumor Tissues and MEG3 Expression Analysis in Cancer Samples
4.3. Cell Line and Culture Condition
4.4. MEG3 Overexpression
4.5. Proliferation Assay
4.6. Clonogenic Assay
4.7. Invasion and Migration Assay
4.8. Spheroid Cultures
4.9. Animals
4.10. Human Tumor Xenograft Growth
4.11. RNA Extraction from Cell Lines and RT-qPCR Analysis
4.12. Western Blot Analysis
4.13. Immunohistochemistry Analysis
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [PubMed] [Green Version]
- Kim, J.; Park, E.Y.; Kim, O.; Schilder, J.M.; Coffey, D.M.; Cho, C.H.; Bast, R.C. Cell origins of high-grade serous ovarian cancer. Cancers 2018, 10, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Auersperg, N. Ovarian Cancers: Their Varied Origins and Pathologically Implicated Microenvironment. In The Ovary, 3rd ed.; Elsevier: London, UK, 2019; Chapter 32; pp. 511–528. [Google Scholar]
- Tang, Y.; Cheung, B.B.; Atmadibrata, B.; Marshall, G.M.; Dinger, M.E.; Liu, P.Y.; Liu, T. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017, 391, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [Green Version]
- Meryet-Figuière, M.; Lambert, B.; Gauduchon, P.; Vigneron, N.; Brotin, E.; Poulain, L.; Denoyelle, C. An overview of long non-coding RNAs in ovarian cancers. Oncotarget 2016, 7, 44719–44734. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Fu, Z.; Dai, C.; Cao, J.; Liu, X.; Xu, J.; Lv, M.; Gu, Y.; Zhang, J.; Hua, X.; et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci. Rep. 2016, 6, 38938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, 45–53. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, X.; Li, J.; Yang, L.; Chen, Z.; Zhao, Q.; Tan, L.; Zhou, Y.; Li, J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol. Rep. 2014, 32, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol. Cancer 2017, 16, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Peng, X.; Dai, Y. The Long Non-Coding RNA (lncRNA) AGAP2-AS1 is Upregulated in Ovarian Carcinoma and Negatively Regulates lncRNA MEG3. Med. Sci. Monit. 2019, 25, 4699–4704. [Google Scholar] [CrossRef] [PubMed]
- Anglesio, M.S.; Wiegand, K.C.; Melnyk, N.; Chow, C.; Salamanca, C.; Prentice, L.M.; Senz, J.; Yang, W.; Spillman, M.A.; Cochrane, D.R.; et al. Type-Specific Cell Line Models for Type-Specific Ovarian Cancer Research. PLoS ONE 2013, 8, e72162. [Google Scholar] [CrossRef]
- Lee, J.M.; Mhawech-Fauceglia, P.; Lee, N.; Parsanian, L.C.; Lin, Y.G.; Gayther, S.A.; Lawrenson, K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Investig. 2013, 93, 528–542. [Google Scholar]
- Beaufort, C.M.; Helmijr, J.C.A.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; Van IJcken, W.F.J.; Heine, A.A.J.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Hallas-Potts, A.; Dawson, J.C.; Herrington, C.S. Ovarian cancer cell lines derived from non-serous carcinomas migrate and invade more aggressively than those derived from high-grade serous carcinomas. Sci. Rep. 2019, 9, 5515. [Google Scholar] [CrossRef]
- Wang, J.; Xu, W.; He, Y.; Xia, Q.; Liu, S. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm. Res. 2018, 67, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Chen, M.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell. Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Filippov-Levy, N.; Cohen-Schussheim, H.; Tropé, C.G.; Hetland Falkenthal, T.E.; Smith, Y.; Davidson, B.; Reich, R. Expression and clinical role of long non-coding RNA in high-grade serous carcinoma. Gynecol. Oncol. 2018, 148, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, M.; Zhao, S. lncRNA MEG3 modified epithelial-mesenchymal transition of ovarian cancer cells by sponging miR-219a-5p and regulating EGFR. J. Cell. Biochem. 2019, 120, 17709–17722. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S. PCAWG Drivers and Functional Interpretation Group; Johnson, R.; PCAWG Consortium. Cancer LncRNA Census reveals evidence fordeep functional conservation of long noncoding RNAs in tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Pulido, T.H.; Guigo, R.; Johnson, R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Luo, Y.; Liang, B.; Ye, L.; Lu, G.; He, W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 2017, 8, 73282–73295. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.A.; Etemadmoghadam, D.; Temple, J.; Lynch, A.G.; Riad, M.; Sharma, R.; Stewart, C.; Fereday, S.; Caldas, C.; Defazio, A.; et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010, 221, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Balas, M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Xu, X.; Li, L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother. Pharmacol. 2017, 79, 479–487. [Google Scholar] [CrossRef]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, F.C.; Santiago, I.; Trinh, A.; Xian, J.; Guo, A.; Sayal, K.; Jimenez-Linan, M.; Deen, S.; Driver, K.; Mack, M.; et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014, 15, 526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wang, C.; Jin, S.; Qu, D.; Ying, H. Expression of NF-kappaB and PTEN in primary epithelial ovarian carcinoma and the correlation with chemoresistance. Int. J. Clin. Exp. Pathol. 2015, 8, 10953–10963. [Google Scholar] [PubMed]
- Dean, M.; Jin, V.; Bergsten, T.M.; Austin, R.; Lantvit, D.D.; Russo, A.; Burdette, J.E. Loss of PTEN in Fallopian Tube Epithelium Results in Multicellular Tumor Spheroid Formation and Metastasis to the Ovary. Cancers 2019, 11, 884. [Google Scholar] [CrossRef] [Green Version]
- Fagotti, A.; Ferrandina, G.; Vizzielli, G.; Fanfani, F.; Gallotta, V.; Chiantera, V.; Costantini, B.; Margariti, P.A.; Gueli Alletti, S.; Cosentino, F.; et al. Phase III randomised clinical trial comparing primary surgery versus neoadjuvant chemotherapy in advanced epithelial ovarian cancer with high tumour load (SCORPION trial): Final analysis of peri-operative outcome. Eur. J. Cancer 2016, 59, 22–33. [Google Scholar] [CrossRef]
- Fagotti, A.; Vizzielli, G.; Ferrandina, G.; Fanfani, F.; Gallotta, V.; Chiantera, V.; Costantini, B.; Margariti, P.A.; Gueli Alletti, S.; Cosentino, F.; et al. Survival analyses from a randomized trial of primary debulking surgery versus neoadjuvant chemotherapy for advanced epithelial ovarian cancer with high tumor load (SCORPION trial). J. Clin. Oncol. 2018, 36, 5516. [Google Scholar] [CrossRef]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; Van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Rustin, G.J.S.; Marples, M.; Nelstrop, A.E.; Mahmoudi, M.; Meyer, T. Use of CA-125 to define progression of ovarian cancer in patients with persistently elevated levels. J. Clin. Oncol. 2001, 19, 4054–4057. [Google Scholar] [CrossRef]
- Colombo, P.E.; Fabbro, M.; Theillet, C.; Bibeau, F.; Rouanet, P.; Ray-Coquard, I. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Crit. Rev. Oncol. Hematol. 2014, 89, 207–216. [Google Scholar] [CrossRef]
- Gallo, D.; Ferlini, C.; Fabrizi, M.; Prislei, S.; Scambia, G. Lack of stimulatory activity of a Phytoestrogen-containing soy extract on the growth of breast cancer tumors in mice. Carcinogenesis 2006, 27, 1404–1409. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Prislei, S.; Martinelli, E.; Zannoni, G.F.; Petrillo, M.; Filippetti, F.; Mariani, M.; Mozzetti, S.; Raspaglio, G.; Scambia, G.; Ferlini, C. Role and prognostic significance of the epithelial-mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget 2015, 6, 18966–18979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Workman, P.; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.; Glennie, M.J.; et al. Committee of the National Cancer Research Institute. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer. 2010, 102, 1555–1577. [Google Scholar] [CrossRef] [Green Version]
- Raspaglio, G.; De Maria, I.; Filippetti, F.; Martinelli, E.; Zannoni, G.F.; Prislei, S.; Ferrandina, G.; Shahabi, S.; Scambia, G.; Ferlini, C. HuR regulates β-tubulin isotype expression in ovarian cancer. Cancer Res. 2010, 70, 5891–5900. [Google Scholar] [CrossRef] [Green Version]
- Van Heesch, S.; Van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; De Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef] [Green Version]
- Khudayberdiev, S.A.; Zampa, F.; Rajman, M.; Schratt, G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013, 6, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Gallo, D.; Zannoni, G.F.; De Stefano, I.; Mosca, M.; Ferlini, C.; Mantuano, E.; Scambia, G. Soy phytochemicals decrease nonsmall cell lung cancer growth in female athymic mice. J. Nutr. 2008, 138, 1360–1364. [Google Scholar] [CrossRef] [Green Version]
- Zannoni, G.F.; Prisco, M.G.; Vellone, V.G.; De Stefano, I.; Scambia, G.; Gallo, D. Changes in the expression of oestrogen receptors and E-cadherin as molecular markers of progression from normal epithelium to invasive cancer in elderly patients with vulvar squamous cell carcinoma. Histopathology. 2011, 58, 265–275. [Google Scholar] [CrossRef]
- Schemper, M.; Smith, T.L. A note on quantifying follow-up in studies of failure time. Control. Clin. Trials 1996, 17, 343–346. [Google Scholar] [CrossRef]




| Characteristics | No. of Patients (%) | |
|---|---|---|
| All cases | 90 | |
| Median age, years (range) | 56 (25–74) | |
| Presence of ascites | ||
| No | 22 (24.4) | |
| Yes | 68 (75.6) | |
| FIGO Stage | ||
| IIIC | 78 (86.6) | |
| IVA | 6 (6.7) | |
| IVB | 6 (6.7) | |
| Primary treatment strategy | ||
| PDS | 56 (62.2) | |
| NACT/IDS | 34 (37.8) | |
| Residual tumor after surgery | ||
| 0 cm | 52 (57.8) | |
| ≤1 cm | 9 (10) | |
| >1 cm | 25 (27.8) | |
| Not available | 4 (4.4) | |
| Primary chemotherapy | ||
| Platinum/paclitaxel | 47 (52.2) | |
| Platinum/paclitaxel/bevacizumab | 36 (40) | |
| Platinum-based | 6 (6.7) | |
| Not available | 1 (1.1) | |
| Chemosensitivity | ||
| Sensitive | 56 (62.2) | |
| Resistant | 34 (37.8) | |
| BRCA mutational status | ||
| BRCA wt | 31 (34.5) | |
| BRCA mut | 20 (22.2) | |
| Not available | 39 (43.3) | |
| Outcome and Variables | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| HR (95%CI) | p | HR (95%CI) | p * | ||
| Age (years) | |||||
| ≤56 | |||||
| >56 | 1.2 (0.8–1.9) | 0.4 | - | - | |
| Ascites | |||||
| No | |||||
| Yes | 0.9 (0.6–1.6) | 0.8 | - | - | |
| FIGO stage | |||||
| IIIC | |||||
| IVA–IVB | 1.5 (0.7–3.1) | 0.3 | - | - | |
| Primary treatment strategy | |||||
| PDS | |||||
| NACT/IDS | 1.2 (0.8–2.0) | 0.4 | - | - | |
| Residual tumor after surgery | |||||
| ≤1 cm | |||||
| >1 cm | 1.7 (1.0–2.9) | 0.07 | 1.5 (0.9–2.4) | 0.1 | |
| MEG3 | |||||
| Low expression | |||||
| High expression | 0.4 (0.2–0.7) | 0.0003 | 0.5 (0.3–0.8) | 0.002 | |
| Outcome and Variables | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| HR (95%CI) | p | HR (95%CI) | p * | ||
| Age (years) | |||||
| ≤56 | |||||
| >56 | 1.7 (0.9–3.1) | 0.09 | 1.8 (1.0–3.3) | 0.05 | |
| Ascites | |||||
| No | |||||
| Yes | 0.9 (0.5–1.9) | 0.9 | - | - | |
| FIGO stage | |||||
| IIIC | |||||
| IVA–IVB | 1.9 (0.7–5.2) | 0.2 | - | - | |
| Primary treatment strategy | |||||
| PDS | |||||
| NACT/IDS | 1.2 (0.7–2.3) | 0.5 | - | - | |
| Residual tumor after surgery | |||||
| ≤1 cm | |||||
| >1 cm | 1.4 (0.7–2.8) | 0.3 | - | - | |
| MEG3 | |||||
| Low expression | |||||
| High expression | 0.5 (0.2–0.8) | 0.01 | 0.4 (0.2–0.8) | 0.01 | |
| Characteristics | No. of Patients with Low MEG3 | No. of Patients with High MEG3 | p * | |
|---|---|---|---|---|
| All cases | 45/90 | 45/90 | ||
| Age (years) | ||||
| ≤56 | 29/50 | 21/50 | 0.1 | |
| >56 | 16/40 | 24/40 | ||
| Ascites | ||||
| No | 12/22 | 10/22 | 0.8 | |
| Yes | 33/68 | 35/68 | ||
| FIGO stage | ||||
| IIIC | 37/78 | 41/78 | 0.4 | |
| IVA–IVB | 8/12 | 4/12 | ||
| Primary treatment strategy | ||||
| PDS | 31/56 | 25/56 | 0.3 | |
| NACT/IDS | 14/34 | 20/34 | ||
| Residual tumor after surgery | ||||
| ≤1 cm | 30/61 | 31/61 | 0.8 | |
| >1 cm | 11/25 | 14/25 | ||
| Not available | 4/4 | 0/4 | ||
| Chemosensitivity | ||||
| Sensitive | 23/56 | 33/56 | 0.05 | |
| Resistant | 22/34 | 12/34 | ||
| BRCA mutational status | ||||
| BRCA wt | 15/31 | 16/31 | 0.1 | |
| BRCA mut | 4/20 | 16/20 | ||
| Not available | 26/39 | 13/39 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buttarelli, M.; De Donato, M.; Raspaglio, G.; Babini, G.; Ciucci, A.; Martinelli, E.; Baccaro, P.; Pasciuto, T.; Fagotti, A.; Scambia, G.; et al. Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 966. https://doi.org/10.3390/cancers12040966
Buttarelli M, De Donato M, Raspaglio G, Babini G, Ciucci A, Martinelli E, Baccaro P, Pasciuto T, Fagotti A, Scambia G, et al. Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer. Cancers. 2020; 12(4):966. https://doi.org/10.3390/cancers12040966
Chicago/Turabian StyleButtarelli, Marianna, Marta De Donato, Giuseppina Raspaglio, Gabriele Babini, Alessandra Ciucci, Enrica Martinelli, Pina Baccaro, Tina Pasciuto, Anna Fagotti, Giovanni Scambia, and et al. 2020. "Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer" Cancers 12, no. 4: 966. https://doi.org/10.3390/cancers12040966
APA StyleButtarelli, M., De Donato, M., Raspaglio, G., Babini, G., Ciucci, A., Martinelli, E., Baccaro, P., Pasciuto, T., Fagotti, A., Scambia, G., & Gallo, D. (2020). Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer. Cancers, 12(4), 966. https://doi.org/10.3390/cancers12040966