The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
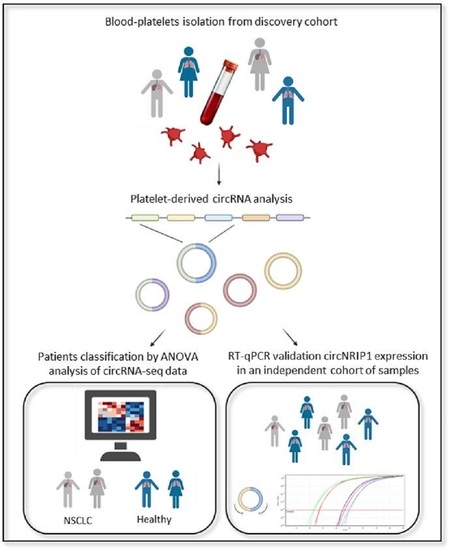
2. Materials and Methods
2.1. Sample Collection and Study Population
2.2. Sample Processing
2.3. RNA Isolation
2.4. cDNA Library Construction
2.5. RNA Sequencing
2.6. RNA Sequencing and Data Processing
2.7. CircRNA Analysis and Identification of DE circRNA
2.8. Reverse Transcription–Quantitative PCR Analysis of circRNAs
3. Results
3.1. Profiling of Platelets circRNA Repertoire by RNA Sequencing and Differential Expression Analysis
3.2. Validation of NRIP1 circRNA by RT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- López-Jiménez, E.; Andres-Leon, E. The Implications of ncRNAs in the Development of Human Disease. Noncoding RNA 2021, 7, 17. [Google Scholar] [PubMed]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, 2345. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157, Erratum in 2013, 19, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2015, 44, 1370–1383. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Nilsson, R.J.; Wurdinger, T. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2014, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak-Wolf, A.; Maier, L.; Mackowiak, S.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Yin, Y.; Long, J.; He, Q.; Li, Y.; Liao, Y.; He, P.; Zhu, W. Emerging roles of circRNA in formation and progression of cancer. J. Cancer 2019, 10, 5015–5021. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Tian, Z.; Fan, W.; Ni, B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 2019, 16, 292–301. [Google Scholar] [CrossRef] [Green Version]
- Debina Sarkar, S.D.D. Circular RNAs: Potential Applications as Therapeutic Targets and Biomarkers in Breast Cancer. Non-Coding RNA 2021, 2, 28. [Google Scholar]
- Micallef, I.; Baron, B. The Mechanistic Roles of ncRNAs in Promoting and Supporting Chemoresistance of Colorectal Cancer. Non-Coding RNA 2021, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, J.; Zhan, X.; Wang, L. Circular RNA Circ-0067934 Attenuates Ferroptosis of Thyroid Cancer Cells by miR-545-3p. Front. Endocrinol. 2021, 12, 1–10. [Google Scholar]
- Wang, Y.; Xu, R.; Zhang, D.; Lu, T.; Yu, W.; Wo, Y.; Liu, A.; Sui, T.; Cui, J.; Qin, Y.; et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl. Lung Cancer Res. 2019, 8, 862–875. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Li, L.; Cai, S.; Zhang, H.; Zhu, X.; Guan, G.; Dong, X. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem. Biophys. Res. Commun. 2018, 503, 2089–2094. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, B.; Wang, J.; Yao, M. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene 2018, 676, 79–85. [Google Scholar] [CrossRef]
- Jin, M.; Shi, C.; Yang, C.; Liu, J.; Huang, G. Upregulated circRNA ARHGAP10 Predicts an Unfavorable Prognosis in NSCLC through Regulation of the miR-150-5p/GLUT-1 Axis. Mol. Ther. Nucleic Acids 2019, 18, 219–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, G.; Zhou, T.; Gu, W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell 2020, 5, 1–36. [Google Scholar] [CrossRef]
- Wang, M.; Xie, F.; Lin, J.; Zhao, Y.; Zhang, Q.; Liao, Z.; Wei, P. Diagnostic and Prognostic Value of Circulating CircRNAs in Cancer. Front. Med. 2021, 8, 1–14. [Google Scholar] [CrossRef]
- Tan, S.; Chien, C.-S.; Yarmishyn, A.; Wang, X. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018, 28, 693–695. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-X.; Yang, Y.-E.; Zhang, M.-Y.; Li, R.; Yin, Y.-H.; Qu, Y.-Q. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 2019, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, D.; Zhou, J.; Qin, N.; Zhou, W.; Ma, H.; Jin, G.; Hu, Z.; Dai, J.; Shen, H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018, 7, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Wei, S.; Chen, Y.; Fan, X.; Han, S.; Wu, G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017, 284, 2170–2182. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-H.; Yang, Y.-P.; Chien, C.-S.; Yarmishyn, A.; Ishola, A.; Chien, Y.; Chen, Y.-M.; Huang, T.-W.; Lee, K.-Y.; Huang, W.-C.; et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers 2020, 12, 1740. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Vancura, A.; Wurdinger, T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J. Thromb. Haemost. 2017, 15, 1295–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Ambrosi, S.; Nilsson, R.J.; Wurdinger, T. Platelets and tumor-associated RNA transfer. Blood 2021, 137, 3181–3191. [Google Scholar] [CrossRef]
- Neu, C.T.; Gutschner, T.; Haemmerle, M. Post-Transcriptional Expression Control in Platelet Biogenesis and Function. Int. J. Mol. Sci. 2020, 110, 7614. [Google Scholar] [CrossRef]
- Gutmann, C.; Zampetaki, A.; Mayr, M. The Landscape of Coding and Non-coding RNAs in Platelets. Antioxid. Redox Signal. 2020, 4, 1–53. [Google Scholar]
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef]
- Best, M.G.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [Green Version]
- Best, M.G.; Sol, N.; Veld, S.G.I.; Vancura, A.; Muller, M.; Niemeijer, A.-L.N.; Fejes, A.V.; Fat, L.-A.T.K.; Veld, A.E.H.I.; Leurs, C.; et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell 2017, 32, 238–252.e9. [Google Scholar] [CrossRef] [PubMed]
- Sol, N.; Veld, S.G.I.; Vancura, A.; Tjerkstra, M.; Leurs, C.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Zwaan, K.; et al. Tumor-Educated Platelet RNA for the Detection and (Pseudo)progression Monitoring of Glioblastoma. Cell Rep. Med. 2020, 1, 101. [Google Scholar] [CrossRef] [PubMed]
- Heinhuis, K.; Veld, S.I.; Dwarshuis, G.; Broek, D.V.D.; Sol, N.; Best, M.; Koenen, A.; Steeghs, N.; Coevorden, F.; Haas, R.; et al. RNA-Sequencing of Tumor-Educated Platelets. A Novel Biomarker for Blood Based Sarcoma Diagnostics. Eur. J. Surg. Oncol. 2020, 46, e7. [Google Scholar] [CrossRef]
- Alhasan, A.A.; Izuogu, O.G.; Al-Balool, H.H.; Steyn, J.S.; Evans, A.; Colzani, M.; Ghevaert, C.; Mountford, J.C.; Marenah, L.; Elliott, D.; et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016, 127, e1–e11. [Google Scholar] [CrossRef]
- Preußer, C.; Hung, L.-H.; Schneider, T.; Schreiner, S.; Hardt, M.; Moebus, A.; Santoso, S.; Bindereif, A. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1424473. [Google Scholar] [CrossRef] [Green Version]
- Stencel, K.; Chmielewska, I.; Milanowski, J.; Ramlau, R. Non-Small-Cell Lung Cancer: New Rare Targets—New Targeted Therapies—State of The Art and Future Directions. Cancers 2021, 13, 1829. [Google Scholar] [CrossRef]
- Sean, T.H.; Sean, C.D.; Knight, B.; Crosbie, P.A.; Balata, H.; Chudziak, J. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar]
- Best, M.G.; In’t Veld, S.G.J.G.; Sol, N.; Wurdinger, T. RNA sequencing and swarm intelligence—Enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat. Protoc. 2019, 14, 1206. [Google Scholar] [CrossRef]
- Sabrkhany, S.; Kuijpers, M.J.; Verheul, H.M.; Egbrink, M.G.O.; Griffioen, A.W. Optimal Human Blood Sampling for Platelet Research. Curr. Angiogenesis 2013, 2, 157–161. [Google Scholar] [CrossRef]
- Sabrkhany, S.; Kuijpers, M.J.; van Kuijk, S.M.; Sanders, L.; Pineda, S.; Damink, S.O.; Dingemans, A.-M.C.; Griffioen, A.W.; Egbrink, M.G.O. A combination of platelet features allows detection of early-stage cancer. Eur. J. Cancer 2017, 80, 5–13. [Google Scholar] [CrossRef]
- Oudejans, C.; Manders, V.; Visser, A.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J.; Berkmortel, T.V.D.; Wigman, D.-J.; Blanken, B.; et al. Circular RNA Sequencing of Maternal Platelets: A Novel Tool for the Identification of Pregnancy-Specific Biomarkers. Clin. Chem. 2021, 67, 508–517. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Conrad, T. Acfs: Accurate circRNA identification and quantification from RNA-Seq data. Sci. Rep. 2016, 6, 38820. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.J.A.; Balaj, L.; Hulleman, E.; Van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.; Vandertop, W.P.; et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Nilsson, R.J.A.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016, 7, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Mohamad, M.A.; Manzor, N.F.M.; Zulkifli, N.F.; Zainal, N.; Hayati, A.R.; Asnawi, A.W.A. A Review of Candidate Genes and Pathways in Preeclampsia—An Integrated Bioinformatical Analysis. Biology 2020, 9, 62. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.R.; Chinnaiyan, A.M. The Potential of Circular RNAS as Cancer Biomarkers. Epidemiol. Biomark. Prev. 2020, 29, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Manders, V.; Visser, A.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J.; Berkmortel, T.V.D.; Hortensius, M.; Jongejan, A.; Pajkrt, E.; et al. The bivariate NRIP1/ZEB2 RNA marker permits non-invasive presymptomatic screening of pre-eclampsia. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Li, X.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis. 2020, 11, 2782. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cai, J.; Han, X.; Ren, Y. Downregulation of circNRIP1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the miR-211-5p/HOXC8 Axis. Cancer Manag. Res. 2020, 12, 9159–9171. [Google Scholar] [CrossRef]
- Li, T.; Shi, C.; Yang, C. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. 2018, 96, 85–96. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, Y.; Xu, L.; Qu, C.; Zhang, L.; Xiao, X.; Chen, W.; Li, K.; Liang, Q.; Wu, H. circ-NRIP1 Promotes Glycolysis and Tumor Progression by Regulating miR-186-5p/MYH9 Axis in Gastric Cancer. Cancer Manag. Res. 2020, 12, 5945–5956. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2021, 40, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Li, M.; Wu, J.; Qin, C.; Tao, Y.; He, H. Circular RNA circNRIP1 Sponges MicroRNA-138-5p to Maintain Hypoxia-Induced Resistance to 5-Fluorouracil Through HIF-1α-Dependent Glucose Metabolism in Gastric Carcinoma. Cancer Manag. Res. 2020, 12, 2789–2802. [Google Scholar] [CrossRef] [Green Version]
- Buzás, E.I.; Tóth, E.; Sódar, B.W.; Szabó-Taylor, K. Molecular interactions at the surface of extracellular vesicles. Semin. Immunopathol. 2018, 40, 453–464. [Google Scholar] [CrossRef] [Green Version]


| N° of Samples | Control | NSCLC |
|---|---|---|
| 6 | 6 | |
| Median age (Min–Max) (years) | 68 (63–80) | 66 (56–80) |
| F/M (%) | 67/33 | 67/33 |
| Localized (%) | - | 33 |
| Metastasized (%) | - | 67 |
| Hospital #1 | 3 | 3 |
| Hospital #2 | 3 | 3 |
| N° of Samples | Control | NSCLC |
|---|---|---|
| 24 | 23 | |
| Median age (Min–Max) (years) | 62.9(51–79) | 60.2 (41–78) |
| F/M (%) | 42/58 | 39/61 |
| Localized (%) | - | 48 |
| Metastasized (%) | - | 52 |
| Hospital #1 | 24 | 11 |
| Hospital #2 | 0 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Ambrosi, S.; Visser, A.; Antunes-Ferreira, M.; Poutsma, A.; Giannoukakos, S.; Sol, N.; Sabrkhany, S.; Bahce, I.; Kuijpers, M.J.E.; Oude Egbrink, M.G.A.; et al. The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers 2021, 13, 4644. https://doi.org/10.3390/cancers13184644
D’Ambrosi S, Visser A, Antunes-Ferreira M, Poutsma A, Giannoukakos S, Sol N, Sabrkhany S, Bahce I, Kuijpers MJE, Oude Egbrink MGA, et al. The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers. 2021; 13(18):4644. https://doi.org/10.3390/cancers13184644
Chicago/Turabian StyleD’Ambrosi, Silvia, Allerdien Visser, Mafalda Antunes-Ferreira, Ankie Poutsma, Stavros Giannoukakos, Nik Sol, Siamack Sabrkhany, Idris Bahce, Marijke J. E. Kuijpers, Mirjam G. A. Oude Egbrink, and et al. 2021. "The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer" Cancers 13, no. 18: 4644. https://doi.org/10.3390/cancers13184644
APA StyleD’Ambrosi, S., Visser, A., Antunes-Ferreira, M., Poutsma, A., Giannoukakos, S., Sol, N., Sabrkhany, S., Bahce, I., Kuijpers, M. J. E., Oude Egbrink, M. G. A., Griffioen, A. W., Best, M. G., Koppers-Lalic, D., Oudejans, C., & Würdinger, T. (2021). The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers, 13(18), 4644. https://doi.org/10.3390/cancers13184644