Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas
Abstract
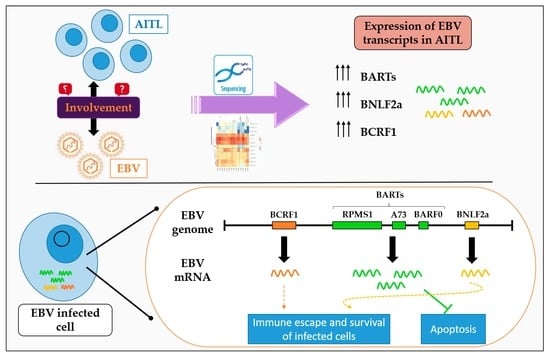
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Analysis of EBV Gene Expression by New Generation Sequencing (NGS)
2.2. AITL Samples Are Not Homogeneous
2.3. BARTs Are the Most Abundant Latency Transcripts in Lymphoma Samples
2.4. All AITL Samples Tested Are in Latency II but Show Strong Expression BNLF2a
2.5. Cell Line Sequencing
3. Discussion
4. Materials and Methods
4.1. Production of Spontaneous LCLs
4.2. Cell Lines and Culture
4.3. Patients
4.4. EBER In Situ Hybridization
4.5. EBV Typing
4.6. RNA Extraction
4.7. mRNA Enrichment
4.8. Probe Design for EBV Sequence Capture
4.9. Sequencing by NGS
4.10. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGeoch, D.J.; Gatherer, D. Lineage Structures in the Genome Sequences of Three Epstein-Barr Virus Strains. Virology 2007, 359, 1–5. [Google Scholar] [CrossRef]
- Smatti, M.K.; Al-Sadeq, D.W.; Ali, N.H.; Pintus, G.; Abou-Saleh, H.; Nasrallah, G.K. Epstein–Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longnecker, R.; Neipel, F. Introduction to the human γ-herpesviruses. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007; ISBN 978-0-521-82714-0. [Google Scholar]
- Wang, C.; Li, D.; Zhang, L.; Jiang, S.; Liang, J.; Narita, Y.; Hou, I.; Zhong, Q.; Zheng, Z.; Xiao, H.; et al. RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, I.A.; Tierney, R.J.; Kelly, G.L.; Stylianou, J.; Rickinson, A.B.; Bell, A.I. Methylation Status of the Epstein-Barr Virus (EBV) BamHI W Latent Cycle Promoter and Promoter Activity: Analysis with Novel EBV-Positive Burkitt and Lymphoblastoid Cell Lines. J. Virol. 2006, 80, 10700–10711. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.-S.; Kieff, E. Epstein-Barr Virus Latent Genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques-Piubelli, M.L.; Salas, Y.I.; Pachas, C.; Becker-Hecker, R.; Vega, F.; Miranda, R.N. Epstein–Barr Virus-Associated B-Cell Lymphoproliferative Disorders and Lymphomas: A Review. Pathology 2020, 52, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, L.S.; Arrand, J.R.; Murray, P.G. EBV gene expression and regulation. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007; ISBN 978-0-521-82714-0. [Google Scholar]
- Grywalska, E.; Rolinski, J. Epstein-Barr Virus-Associated Lymphomas. Semin. Oncol. 2015, 42, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Rasul, A.E.; Nagy, N.; Sohlberg, E.; Ádori, M.; Claesson, H.-E.; Klein, G.; Klein, E. Simultaneous Detection of the Two Main Proliferation Driving EBV Encoded Proteins, EBNA-2 and LMP-1 in Single B Cells. J. Immunol. Methods 2012, 385, 60–70. [Google Scholar] [CrossRef]
- Klein, E.; Nagy, N.; Rasul, A.E. EBV Genome Carrying B Lymphocytes That Express the Nuclear Protein EBNA-2 but Not LMP-1: Type IIb Latency. Oncoimmunology 2013, 2, e23035. [Google Scholar] [CrossRef] [Green Version]
- Price, A.M.; Luftig, M.A. To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef] [PubMed]
- Messinger, J.E.; Dai, J.; Stanland, L.J.; Price, A.M.; Luftig, M.A. Identification of Host Biomarkers of Epstein-Barr Virus Latency IIb and Latency III. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenzie, J.; El-Guindy, A. Epstein-Barr Virus Lytic Cycle Reactivation. Curr. Top. Microbiol. Immunol. 2015, 391, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Concha, M.; Wang, X.; Cao, S.; Baddoo, M.; Fewell, C.; Lin, Z.; Hulme, W.; Hedges, D.; McBride, J.; Flemington, E.K. Identification of New Viral Genes and Transcript Isoforms during Epstein-Barr Virus Reactivation Using RNA-Seq. J. Virol. 2012, 86, 1458–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivachandran, N.; Wang, X.; Frappier, L. Functions of the Epstein-Barr Virus EBNA1 Protein in Viral Reactivation and Lytic Infection. J. Virol. 2012, 86, 6146–6158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravorty, S.; Yan, B.; Wang, C.; Wang, L.; Quaid, J.T.; Lin, C.F.; Briggs, S.D.; Majumder, J.; Canaria, D.A.; Chauss, D.; et al. Integrated Pan-Cancer Map of EBV-Associated Neoplasms Reveals Functional Host-Virus Interactions. Cancer Res. 2019, 79, 6010–6023. [Google Scholar] [CrossRef] [Green Version]
- Shannon-Lowe, C.; Rickinson, A. The Global Landscape of EBV-Associated Tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef] [Green Version]
- De Leval, L.; Parrens, M.; Le Bras, F.; Jais, J.-P.; Fataccioli, V.; Martin, A.; Lamant, L.; Delarue, R.; Berger, F.; Arbion, F.; et al. Angioimmunoblastic T-Cell Lymphoma Is the Most Common T-Cell Lymphoma in Two Distinct French Information Data Sets. Haematologica 2015, 100, e361–e364. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Sakata-Yanagimoto, M. Advances in Understanding of Angioimmunoblastic T-Cell Lymphoma. Leukemia 2020, 34, 2592–2606. [Google Scholar] [CrossRef]
- Vose, J.; Armitage, J.; Weisenburger, D. International T-Cell Lymphoma Project International Peripheral T-Cell and Natural Killer/T-Cell Lymphoma Study: Pathology Findings and Clinical Outcomes. J. Clin. Oncol. 2008, 26, 4124–4130. [Google Scholar] [CrossRef]
- Federico, M.; Rudiger, T.; Bellei, M.; Nathwani, B.N.; Luminari, S.; Coiffier, B.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Savage, K.J.; et al. Clinicopathologic Characteristics of Angioimmunoblastic T-Cell Lymphoma: Analysis of the International Peripheral T-Cell Lymphoma Project. J. Clin. Oncol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [Green Version]
- Anagnostopoulos, I.; Hummel, M.; Finn, T.; Tiemann, M.; Korbjuhn, P.; Dimmler, C.; Gatter, K.; Dallenbach, F.; Parwaresch, M.R.; Stein, H. Heterogeneous Epstein-Barr Virus Infection Patterns in Peripheral T-Cell Lymphoma of Angioimmunoblastic Lymphadenopathy Type. Blood 1992, 80, 1804–1812. [Google Scholar] [CrossRef] [Green Version]
- Beer, T.; Dorion, P. Angioimmunoblastic T-Cell Lymphoma Presenting with an Acute Serologic Epstein-Barr Virus Profile. Hematol. Rep. 2015, 7, 5893. [Google Scholar] [CrossRef] [Green Version]
- Willenbrock, K.; Bräuninger, A.; Hansmann, M.-L. Frequent Occurrence of B-Cell Lymphomas in Angioimmunoblastic T-Cell Lymphoma and Proliferation of Epstein-Barr Virus-Infected Cells in Early Cases. Br. J. Haematol. 2007, 138, 733–739. [Google Scholar] [CrossRef]
- Xu, Y.; McKenna, R.W.; Hoang, M.P.; Collins, R.H.; Kroft, S.H. Composite Angioimmunoblastic T-Cell Lymphoma and Diffuse Large B-Cell Lymphoma: A Case Report and Review of the Literature. Am. J. Clin. Pathol. 2002, 118, 848–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zettl, A.; Lee, S.-S.; Rüdiger, T.; Starostik, P.; Marino, M.; Kirchner, T.; Ott, M.; Müller-Hermelink, H.K.; Ott, G. Epstein-Barr Virus-Associated B-Cell Lymphoproliferative Disorders in Angloimmunoblastic T-Cell Lymphoma and Peripheral T-Cell Lymphoma, Unspecified. Am. J. Clin. Pathol. 2002, 117, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, K.; Wilson, W.H. Angioimmunoblastic T-Cell Lymphoma: Immune Modulation as a Therapeutic Strategy. Leuk. Lymph. 2007, 48, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Nakhoul, H.; Lin, Z.; Wang, X.; Roberts, C.; Dong, Y.; Flemington, E. High-Throughput Sequence Analysis of Peripheral T-Cell Lymphomas Indicates Subtype-Specific Viral Gene Expression Patterns and Immune Cell Microenvironments. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.R.; de Jesus, O.; Turner, D.; Hollyoake, M.; Karstegl, C.E.; Griffin, B.E.; Karran, L.; Wang, Y.; Hayward, S.D.; Farrell, P.J. Structure and Coding Content of CST (BART) Family RNAs of Epstein-Barr Virus. J. Virol. 2000, 74, 3082–3092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skare, J.; Edson, C.; Farley, J.; Strominger, J.L. The B95-8 Isolate of Epstein-Barr Virus Arose from an Isolate with a Standard Genome. J. Virol. 1982, 44, 1088–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Lin, Z.; Wu, Y.; Dong, J.; Zhao, B.; Cheng, Y.; Huang, P.; Xu, L.; Xia, T.; Xiong, D.; et al. Comprehensive Profiling of EBV Gene Expression in Nasopharyngeal Carcinoma through Paired-End Transcriptome Sequencing. Front. Med. 2016, 10, 61–75. [Google Scholar] [CrossRef]
- Calderwood, M.A.; Holthaus, A.M.; Johannsen, E. The Epstein-Barr Virus LF2 Protein Inhibits Viral Replication. J. Virol. 2008, 82, 8509–8519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, B.D.; Bankier, A.; Satchwell, S.; Barrell, B.; Farrell, P.J. Sequence and Transcription of Raji Epstein-Barr Virus DNA Spanning the B95-8 Deletion Region. Virology. 1990, 179, 339–346. [Google Scholar] [CrossRef]
- Southam, C.M.; Burchenal, J.H.; Clarkson, B.; Tanzi, A.; Mackey, R.; McComb, V. Hetero- Transplantation of Human Cell Lines from Burkitt’s Tumors and Acute Leukemia into Newborn Rats. Cancer 1969, 23, 281–299. [Google Scholar] [CrossRef]
- Greijer, A.E.; Ramayanti, O.; Verkuijlen, S.a.W.M.; Novalić, Z.; Juwana, H.; Middeldorp, J.M. Quantitative Multi-Target RNA Profiling in Epstein-Barr Virus Infected Tumor Cells. J. Virol. Methods 2017, 241, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Dombos, L.; Gothoskar, B. Sensitivity of Epstein-Barr Virus (EBV) Producer and Non-Producer Human Lymphoblastoid Cell Lines to Superinfection with EB-Virus. Int. J. Cancer. 1972, 10, 44–57. [Google Scholar] [CrossRef]
- Bernasconi, M.; Berger, C.; Sigrist, J.A.; Bonanomi, A.; Sobek, J.; Niggli, F.K.; Nadal, D. Quantitative Profiling of Housekeeping and Epstein-Barr Virus Gene Transcription in Burkitt Lymphoma Cell Lines Using an Oligonucleotide Microarray. Virol. J. 2006, 3, 43. [Google Scholar] [CrossRef] [Green Version]
- Henderson, A.; Ripley, S.; Heller, M.; Kieff, E. Chromosome Site for Epstein-Barr Virus DNA in a Burkitt Tumor Cell Line and in Lymphocytes Growth-Transformed in Vitro. Proc. Natl. Acad. Sci. USA 1983, 80, 1987–1991. [Google Scholar] [CrossRef] [Green Version]
- Hinuma, Y.; Konn, M.; Yamaguchi, J.; Wudarski, D.J.; Blakeslee, J.R.; Grace, J.T. Immunofluorescence and Herpes-Type Virus Particles in the P3HR-1 Burkitt Lymphoma Cell Line. J. Virol. 1967, 1, 1045–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawada, J.-I.; Ando, S.; Torii, Y.; Watanabe, T.; Sato, Y.; Ito, Y.; Kimura, H. Antitumor Effects of Duvelisib on Epstein-Barr Virus-Associated Lymphoma Cells. Cancer Med. 2018, 7, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Chelouah, S.; Cochet, E.; Couvé, S.; Balkaran, S.; Robert, A.; May, E.; Ogryzko, V.; Wiels, J. New Interactors of the Truncated EBNA-LP Protein Identified by Mass Spectrometry in P3HR1 Burkitt’s Lymphoma Cells. Cancers 2018, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Young, L.S.; Rickinson, A.B. Epstein-Barr Virus: 40 Years On. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M.; Zajac, B.; Henle, G.; Henle, W. Morphological and Virological Investigations on Cultured Burkitt Tumor Lymphoblasts (Strain Raji). J. Natl. Cancer Inst. 1966, 37, 547–559. [Google Scholar]
- Decaussin, G.; Leclerc, V.; Ooka, T. The Lytic Cycle of Epstein-Barr Virus in the Nonproducer Raji Line Can Be Rescued by the Expression of a 135-Kilodalton Protein Encoded by the BALF2 Open Reading Frame. J. Virol. 1995, 69, 7309–7314. [Google Scholar] [CrossRef] [Green Version]
- Hatfull, G.; Bankier, A.T.; Barrell, B.G.; Farrell, P.J. Sequence Analysis of Raji Epstein-Barr Virus DNA. Virology 1988, 164, 334–340. [Google Scholar] [CrossRef]
- Coppo, P.; Gouilleux-Gruart, V.; Huang, Y.; Bouhlal, H.; Bouamar, H.; Bouchet, S.; Perrot, C.; Vieillard, V.; Dartigues, P.; Gaulard, P.; et al. STAT3 Transcription Factor Is Constitutively Activated and Is Oncogenic in Nasal-Type NK/T-Cell Lymphoma. Leukemia 2009, 23, 1667–1678. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Nagata, H.; Ikeuchi, T.; Mukai, H.; Oyoshi, M.K.; Demachi, A.; Morio, T.; Wakiguchi, H.; Kimura, N.; Shimizu, N.; et al. Common Cytological and Cytogenetic Features of Epstein-Barr Virus (EBV)-Positive Natural Killer (NK) Cells and Cell Lines Derived from Patients with Nasal T/NK-Cell Lymphomas, Chronic Active EBV Infection and Hydroa Vacciniforme-like Eruptions. Br. J. Haematol. 2003, 121, 805–814. [Google Scholar] [CrossRef]
- Takahara, M.; Kis, L.L.; Nagy, N.; Liu, A.; Harabuchi, Y.; Klein, G.; Klein, E. Concomitant Increase of LMP1 and CD25 (IL-2-Receptor Alpha) Expression Induced by IL-10 in the EBV-Positive NK Lines SNK6 and KAI3. Int. J. Cancer 2006, 119, 2775–2783. [Google Scholar] [CrossRef]
- Ishii, H.; Takahara, M.; Nagato, T.; Kis, L.L.; Nagy, N.; Kishibe, K.; Harabuchi, Y.; Klein, E. Monocytes Enhance Cell Proliferation and LMP1 Expression of Nasal Natural Killer/T-Cell Lymphoma Cells by Cell Contact-Dependent Interaction through Membrane-Bound IL-15. Int. J. Cancer 2012, 130, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Hitt, M.M.; Allday, M.J.; Hara, T.; Karran, L.; Jones, M.D.; Busson, P.; Tursz, T.; Ernberg, I.; Griffin, B.E. EBV Gene Expression in an NPC-Related Tumour. EMBO J. 1989, 8, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, K.J.; Rajadurai, P.; Lin, J.C.; Busson, P.; Abdel-Hamid, M.; Prasad, U.; Tursz, T.; Raab-Traub, N. Expression of the Epstein-Barr Virus BamHI a Fragment in Nasopharyngeal Carcinoma: Evidence for a Viral Protein Expressed in Vivo. J. Virol. 1991, 65, 6252–6259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dambaugh, T.; Nkrumah, F.K.; Biggar, R.J.; Kieff, E. Epstein-Barr Virus RNA in Burkitt Tumor Tissue. Cell 1979, 16, 313–322. [Google Scholar] [CrossRef]
- Verhoeven, R.J.A.; Tong, S.; Mok, B.W.-Y.; Liu, J.; He, S.; Zong, J.; Chen, Y.; Tsao, S.-W.; Lung, M.L.; Chen, H. Epstein-Barr Virus BART Long Non-Coding RNAs Function as Epigenetic Modulators in Nasopharyngeal Carcinoma. Front Oncol 2019, 9, 1120. [Google Scholar] [CrossRef] [Green Version]
- Strong, M.J.; Xu, G.; Coco, J.; Baribault, C.; Vinay, D.S.; Lacey, M.R.; Strong, A.L.; Lehman, T.A.; Seddon, M.B.; Lin, Z.; et al. Differences in Gastric Carcinoma Microenvironment Stratify According to EBV Infection Intensity: Implications for Possible Immune Adjuvant Therapy. PLoS Pathog. 2013, 9, e1003341. [Google Scholar] [CrossRef]
- Marquitz, A.R.; Mathur, A.; Edwards, R.H.; Raab-Traub, N. Host Gene Expression Is Regulated by Two Types of Noncoding RNAs Transcribed from the Epstein-Barr Virus BamHI a Rightward Transcript Region. J. Virol. 2015, 89, 11256–11268. [Google Scholar] [CrossRef] [Green Version]
- Marquitz, A.R.; Raab-Traub, N. The Role of MiRNAs and EBV BARTs in NPC. Semin. Cancer Biol. 2012, 22, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Al-Mozaini, M.; Bodelon, G.; Karstegl, C.E.; Jin, B.; Al-Ahdal, M.; Farrell, P.J. Epstein-Barr Virus BART Gene Expression. J. Gen. Virol. 2009, 90, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Schäfer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein-Barr Virus MicroRNAs Are Evolutionarily Conserved and Differentially Expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.Y.; Pfuhl, T.; Motsch, N.; Barth, S.; Nicholls, J.; Grässer, F.; Meister, G. Identification of Novel Epstein-Barr Virus MicroRNA Genes from Nasopharyngeal Carcinomas. J. Virol. 2009, 83, 3333–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, K.; Yokoyama, N.; Kiyono, T.; Kuzushima, K.; Homma, M.; Nishiyama, Y.; Fujita, M.; Tsurumi, T. The Epstein-Barr Virus Pol Catalytic Subunit Physically Interacts with the BBLF4-BSLF1-BBLF2/3 Complex. J. Virol. 2000, 74, 2550–2557. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Lim, Y.; Im, H.; Bae, J.M.; Kang, G.H.; Ahn, J.; Baek, D.; Kim, T.-Y.; Yoon, S.-S.; Koh, Y. Interpretation of EBV Infection in Pan-Cancer Genome Considering Viral Life Cycle: LiEB (Life Cycle of Epstein-Barr Virus). Sci. Rep. 2019, 9, 3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kheir, F.; Zhao, M.; Strong, M.J.; Yu, Y.; Nanbo, A.; Flemington, E.K.; Morris, G.F.; Reiss, K.; Li, L.; Lin, Z. Detection of Epstein-Barr Virus Infection in Non-Small Cell Lung Cancer. Cancers 2019, 11, 759. [Google Scholar] [CrossRef] [Green Version]
- Strong, M.J.; Laskow, T.; Nakhoul, H.; Blanchard, E.; Liu, Y.; Wang, X.; Baddoo, M.; Lin, Z.; Yin, Q.; Flemington, E.K. Latent Expression of the Epstein-Barr Virus (EBV)-Encoded Major Histocompatibility Complex Class I TAP Inhibitor, BNLF2a, in EBV-Positive Gastric Carcinomas. J. Virol. 2015, 89, 10110–10114. [Google Scholar] [CrossRef] [Green Version]
- Jochum, S.; Moosmann, A.; Lang, S.; Hammerschmidt, W.; Zeidler, R. The EBV Immunoevasins VIL-10 and BNLF2a Protect Newly Infected B Cells from Immune Recognition and Elimination. PLoS Pathog. 2012, 8, e1002704. [Google Scholar] [CrossRef] [Green Version]
- Horst, D.; van Leeuwen, D.; Croft, N.P.; Garstka, M.A.; Hislop, A.D.; Kremmer, E.; Rickinson, A.B.; Wiertz, E.J.H.J.; Ressing, M.E. Specific Targeting of the EBV Lytic Phase Protein BNLF2a to the Transporter Associated with Antigen Processing Results in Impairment of HLA Class I-Restricted Antigen Presentation. J. Immunol. 2009, 182, 2313–2324. [Google Scholar] [CrossRef] [Green Version]
- Croft, N.P.; Shannon-Lowe, C.; Bell, A.I.; Horst, D.; Kremmer, E.; Ressing, M.E.; Wiertz, E.J.H.J.; Middeldorp, J.M.; Rowe, M.; Rickinson, A.B.; et al. Stage-Specific Inhibition of MHC Class I Presentation by the Epstein-Barr Virus BNLF2a Protein during Virus Lytic Cycle. PLoS Pathog. 2009, 5, e1000490. [Google Scholar] [CrossRef]
- Rojas, J.M.; Avia, M.; Martín, V.; Sevilla, N. IL-10: A Multifunctional Cytokine in Viral Infections. J. Immunol. Res. 2017, 2017, 6104054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jog, N.R.; Chakravarty, E.F.; Guthridge, J.M.; James, J.A. Epstein Barr Virus Interleukin 10 Suppresses Anti-Inflammatory Phenotype in Human Monocytes. Front. Immunol. 2018, 9, 2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borozan, I.; Zapatka, M.; Frappier, L.; Ferretti, V. Analysis of Epstein-Barr Virus Genomes and Expression Profiles in Gastric Adenocarcinoma. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooykaas, M.J.G.; Kruse, E.; Wiertz, E.J.H.J.; Lebbink, R.J. Comprehensive Profiling of Functional Epstein-Barr Virus MiRNA Expression in Human Cell Lines. BMC Genom. 2016, 17, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portis, T.; Longnecker, R. Epstein–Barr Virus (EBV) LMP2A Mediates B-Lymphocyte Survival through Constitutive Activation of the Ras/PI3K/Akt Pathway. Oncogene 2004, 23, 8619–8628. [Google Scholar] [CrossRef] [Green Version]
- Incrocci, R.; Barse, L.; Stone, A.; Vagvala, S.; Montesano, M.; Subramaniam, V.; Swanson-Mungerson, M. Epstein-Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances IL-10 Production through the Activation of Bruton’s Tyrosine Kinase and STAT3. Virology 2017, 500, 96–102. [Google Scholar] [CrossRef]
- Henderson, S.; Huen, D.; Rowe, M.; Dawson, C.; Johnson, G.; Rickinson, A. Epstein-Barr Virus-Coded BHRF1 Protein, a Viral Homologue of Bcl-2, Protects Human B Cells from Programmed Cell Death. Proc. Natl. Acad. Sci. USA 1993, 90, 8479–8483. [Google Scholar] [CrossRef] [Green Version]
- Altmann, M.; Hammerschmidt, W. Epstein-Barr Virus Provides a New Paradigm: A Requirement for the Immediate Inhibition of Apoptosis. PLoS Biol. 2005, 3. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, K.; Haar, J.; Tsai, M.-H.; Poirey, R.; Feederle, R.; Delecluse, H.-J. A Viral MicroRNA Cluster Regulates the Expression of PTEN, P27 and of a Bcl-2 Homolog. PLoS Pathog. 2016, 12, e1005405. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Qu, J.; Peng, Q.; Gan, R. Molecular Mechanisms of EBV-Driven Cell Cycle Progression and Oncogenesis. Med. Microbiol. Immunol. 2019, 208, 573–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, A.G.; McLachlan, S.M.; Britton, S. Cyclosporin A Promotes Spontaneous Outgrowth in Vitro of Epstein-Barr Virus-Induced B-Cell Lines. Nature 1981, 289, 300–301. [Google Scholar] [CrossRef]
- Sculley, T.B.; Moss, D.J.; Hazelton, R.A.; Pope, J.H. Detection of Epstein-Barr Virus Strain Variants in Lymphoblastoid Cell Lines “spontaneously” Derived from Patients with Rheumatoid Arthritis, Infectious Mononucleosis and Normal Controls. J. Gen. Virol. 1987, 68, 2069–2078. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [Green Version]






| Sample Name | Read Number | Mean Depth | EBV Type |
|---|---|---|---|
| B95-8 | 10,053,705 | 2010 | EBV-1 |
| CoAN | 1,942,331 | 1251 | EBV-1 |
| DPL | 3,490,546 | 1326 | EBV-1 |
| Jijoye | 5,538,086 | 1586 | EBV-1 |
| KREB2 | 2,194,143 | 117 | EBV-1 |
| MECO4 | 894,293 | 61 | EBV-2 |
| MLEB2 | 2,351,251 | 537 | EBV-1 |
| Namalwa | 2,697,676 | 778 | EBV-2 |
| P3HR1 | 3,622,454 | 698 | EBV-1 |
| Raji | 1,993,483 | 65 | EBV-1 |
| SNK6 | 3,175,449 | 971 | EBV-2 |
| PTBL5 | 1,033,508 | 63 | EBV-1 |
| DLBCL3 | 1,071,114 | 42 | EBV-1 |
| AIL7 | 1,248,228 | 71 | EBV-1 |
| ARL2 | 3,254,648 | 873 | EBV-1 |
| AIL15 | 2,356,168 | 626 | EBV-1 |
| ALCL1 | 1,782,932 | 78 | EBV-1 |
| AIL25 | 3,143,910 | 751 | EBV-1 |
| AIL24 | 1,204,758 | 56 | EBV-1 |
| AIL27 | 889,930 | 45 | EBV-1 |
| AIL2 | 2,024,732 | 92 | EBV-1 |
| AIL11 | 910,244 | 47 | EBV-1 |
| DLBCL4 | 1,764,060 | 301 | EBV-1 |
| PTBL4 | 1,494,164 | 385 | EBV-1 |
| AIL3 | 1,066,140 | 46 | EBV-1 |
| CTCL1 | 4,930,412 | 792 | EBV-1 |
| PTBL6 | 1,009,426 | 180 | EBV-1 |
| AIL14 | 1,948,266 | 553 | EBV-1 |
| AIL16 | 1,214,934 | 63 | EBV-1 |
| NLPHL2 | 2,009,568 | 45 | EBV-1 |
| NS.CHL1 | 566,500 | 51 | EBV-1 |
| NS.CHL3 | 3,573,262 | 861 | EBV-1 |
| MC.CHL1 | 1,907,892 | 133 | EBV-1 |
| AIL22 | 1,168,146 | 46 | EBV-1 |
| NS.CHL2 | 3,669,640 | 48 | EBV-1 |
| AIL23 | 2,096,084 | 47 | EBV-1 |
| AIL26 | 1,360,172 | 42 | EBV-1 |
| PTCL.NOS4 | 1,752,840 | 45 | EBV-1 |
| AIL21 | 2,526,360 | 102 | EBV-1 |
| ALCL2 | 4,104,334 | 43 | EBV-1 |
| CTCL2 | 1,376,010 | 159 | EBV-1 |
| CTCL3 | 1,495,068 | 124 | EBV-1 |
| PTCL.NOS5 | 2,894,860 | 47 | EBV-1 |
| PTCL.NOS3 | 1,361,598 | 42 | EBV-1 |
| NLPHL1 | 2,610,342 | 74 | EBV-1 |
| DLBCL5 | 8,303,756 | 1027 | EBV-1 |
| Cell Line | Origin | Latency Type | Particularity |
|---|---|---|---|
| B95.8 | Primary infection [31] | Lat III [32] | Deletion 139,724 bp to 151,554 bp: OriLyt—large part of miRNA BART—LF-1, -2, -3 [31,33,34] |
| Jijoye | Endemic Burkitt’s lymphoma [35] | Lat III [36] | No deletion |
| Namalwa | Endemic Burkitt’s lymphoma [37] | Lat I [38] Lat III [36] | 2 copies of EBV genome integrated into the human chromosome [39] |
| P3HR1 | Burkitt’s lymphoma [40] | Lat I [36] Lat II [41] Atypical latency [42] | Derived from Jijoye—Deletion (33,355 bp to 40,163 bp): EBNA-2, part of EBNA-LP, part of BHLF-1 [34,38,43] |
| Raji | Burkitt’s lymphoma [44] | Lat III [36] Lat I/Lat III (in vitro) [38] | Two deletions (99,126 bp to 102,118 bp and 163,978 bp to 166,635 bp): EBNA-3C, BZLF2, BARF1, BALF1, BALF2 [45,46] |
| MEC04 | NK/T lymphoma [47] | Lat II [47] | |
| SNK6 | NK/T lymphoma [48] | Lat II [48] | EBNA-2 not expressed [49,50] |
| Cell Line | Patient Age | Patient Sex | Patient Pathology |
|---|---|---|---|
| CoAN | 61 | F | Renal cell carcinoma and hepatocellular carcinoma |
| DPL | 46 | M | Cardiac AL λ amyloidosis |
| KREB2 | 64 | M | Parsonage-Turner syndrome |
| MLEB2 | 66 | M | Healthy subject |
| Patient | Sex | Age at Diagnosis | Pathology According to WHO Criteria (2016) |
|---|---|---|---|
| AIL2 | M | 62 | Angioimmunoblastic T-cell lymphoma |
| AIL3 | M | 80 | Angioimmunoblastic T-cell lymphoma |
| AIL7 | F | 59 | Angioimmunoblastic T-cell lymphoma |
| AIL11 | M | 59 | Angioimmunoblastic T-cell lymphoma |
| AIL14 | M | 62 | Angioimmunoblastic T-cell lymphoma |
| AIL15 | M | 67 | Angioimmunoblastic T-cell lymphoma |
| AIL16 | M | 50 | Angioimmunoblastic T-cell lymphoma |
| AIL21 | M | 79 | Angioimmunoblastic T-cell lymphoma |
| AIL22 | M | 70 | Angioimmunoblastic T-cell lymphoma |
| AIL23 | M | 67 | Angioimmunoblastic T-cell lymphoma |
| AIL24 | F | 78 | Angioimmunoblastic T-cell lymphoma |
| AIL25 | M | 59 | Angioimmunoblastic T-cell lymphoma |
| AIL26 | M | 69 | Angioimmunoblastic T-cell lymphoma |
| AIL27 | F | 69 | Angioimmunoblastic T-cell lymphoma |
| PTCL-NOS3 | M | 81 | Peripheral T-cell lymphoma, not otherwise specified |
| PTCL-NOS4 | M | 69 | Peripheral T-cell lymphoma, not otherwise specified |
| PTCL-NOS5 | F | 80 | Peripheral T-cell lymphoma, not otherwise specified |
| ALCL1 | F | 73 | Anaplastic large T cell lymphoma |
| ALCL2 | M | 20 | Anaplastic large T cell lymphoma |
| CTCL1 | F | 73 | Cutaneous T cell lymphoma |
| CTCL2 | M | 63 | Cutaneous T cell lymphoma |
| CTCL3 | M | 83 | Cutaneous T cell lymphoma |
| NLPHL1 | M | 68 | Nodular lymphocyte-predominant type Hodgkin’s lymphoma |
| NLPHL2 | M | 33 | Nodular lymphocyte-predominant type Hodgkin’s lymphoma |
| NS-CHL1 | M | 20 | Nodular sclerosis classical Hodgkin’s lymphoma |
| NS-CHL2 | M | 73 | Nodular sclerosis classical Hodgkin’s lymphoma |
| NS-CHL3 | F | 67 | Nodular sclerosis classical Hodgkin’s lymphoma |
| MC-CHL1 | M | 77 | Mixed cellularity classical Hodgkin’s lymphoma |
| DLBCL3 | F | 59 | Diffuse large B-cell lymphoma |
| DLBCL4 | F | 31 | Diffuse large B-cell lymphoma |
| DLBCL5 | F | 59 | Diffuse large B-cell lymphoma |
| PTBL4 | F | 52 | Post-transplant B lymphoma |
| PTBL5 | F | 68 | Post-transplant B lymphoma |
| PTBL6 | M | 57 | Post-transplant B lymphoma |
| ARL2 | F | 76 | Age-related lymphoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayda, N.; Tilloy, V.; Chaunavel, A.; Bahri, R.; Halabi, M.A.; Feuillard, J.; Jaccard, A.; Ranger-Rogez, S. Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas. Cancers 2021, 13, 610. https://doi.org/10.3390/cancers13040610
Bayda N, Tilloy V, Chaunavel A, Bahri R, Halabi MA, Feuillard J, Jaccard A, Ranger-Rogez S. Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas. Cancers. 2021; 13(4):610. https://doi.org/10.3390/cancers13040610
Chicago/Turabian StyleBayda, Nader, Valentin Tilloy, Alain Chaunavel, Racha Bahri, Mohamad Adnan Halabi, Jean Feuillard, Arnaud Jaccard, and Sylvie Ranger-Rogez. 2021. "Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas" Cancers 13, no. 4: 610. https://doi.org/10.3390/cancers13040610
APA StyleBayda, N., Tilloy, V., Chaunavel, A., Bahri, R., Halabi, M. A., Feuillard, J., Jaccard, A., & Ranger-Rogez, S. (2021). Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas. Cancers, 13(4), 610. https://doi.org/10.3390/cancers13040610