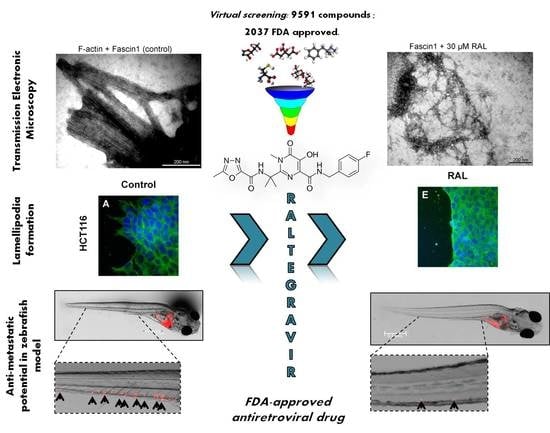
The FDA-Approved Antiviral Raltegravir Inhibits Fascin1-Dependent Invasion of Colorectal Tumor Cells In Vitro and In Vivo
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Virtual Screening
2.2. Molecular Dynamics
2.3. Structural Comparison
2.4. Compound Purchase-Chemistry
2.5. Recombinant Fascin1 Expression and Purification
2.6. Thermofluor and Fluorescence Titration
2.7. Ligand-Observed NMR
2.8. Transmission Electron Microscopy Detection
2.9. Cell Culture
2.10. Cell Viability Assay
2.11. Cell-Migration Assay
2.12. Transwell Invasion Assay
2.13. Wounding-Scratch Assay Immunofluorescence
2.14. Zebrafish Invasion and Metastasis Assays
2.15. Data Analysis
3. Results
3.1. Virtual Screening and Molecular Dynamics
3.2. Differential Scanning Fluorimetry (Thermofluor) Study of Binding to Fascin1
3.3. RAL Prevents In Vitro Fascin1-Induced F-Actin Bundling
3.4. RAL Affects Actin Cytoskeleton Formation and Fascin1 Localization at Lamellipodia
3.5. RAL Diminishes Migration and Inhibits Matrigel Cell Invasion of Colorectal Cancer Cells
3.6. RAL Inhibits the Invasive and Metastatic Capacity of Colorectal Tumor Cells in an In Vivo Model
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A







References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Conesa-Zamora, P.; García-Solano, J.; Turpin, M.e.C.; Sebastián-León, P.; Torres-Moreno, D.; Estrada, E.; Tuomisto, A.; Wilce, J.; Mäkinen, M.J.; Pérez-Guillermo, M.; et al. Methylome profiling reveals functions and genes which are differentially methylated in serrated compared to conventional colorectal carcinoma. Clin. Epigenet. 2015, 7, 101. [Google Scholar] [CrossRef] [Green Version]
- García-Solano, J.; Conesa-Zamora, P.; Carbonell, P.; Trujillo-Santos, J.; Torres-Moreno, D.; Pagán-Gómez, I.; Rodríguez-Braun, E.; Pérez-Guillermo, M. Colorectal serrated adenocarcinoma shows a different profile of oncogene mutations, MSI status and DNA repair protein expression compared to conventional and sporadic MSI-H carcinomas. Int. J. Cancer 2012, 131, 1790–1799. [Google Scholar] [CrossRef]
- Stefanius, K.; Ylitalo, L.; Tuomisto, A.; Kuivila, R.; Kantola, T.; Sirniö, P.; Karttunen, T.J.; Mäkinen, M.J. Frequent mutations of KRAS in addition to BRAF in colorectal serrated adenocarcinoma. Histopathology 2011, 58, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Alburquerque-González, B.; López-Calderón, F.F.; López-Abellán, M.D.; Esteban-Gil, Á.; García-Solano, J.; Conesa-Zamora, P. Biology and Therapeutic Targets of Colorectal Serrated Adenocarcinoma; Clues for a Histologically Based Treatment against an Aggressive Tumor. Int. J. Mol. Sci. 2020, 21, 1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, V.Y.; Lewis, S.J.; Adams, J.C.; Martin, R.M. Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: A systematic review and meta-analysis. BMC Med. 2013, 11, 52. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, P.C.; Sawazaki-Calone, I.; de Oliveira, C.E.; Soares Macedo, C.C.; Dourado, M.R.; Cervigne, N.K.; Miguel, M.C.; Ferreira do Carmo, A.; Lambert, D.W.; Graner, E.; et al. Fascin promotes migration and invasion and is a prognostic marker for oral squamous cell carcinoma. Oncotarget 2017, 8, 74736–74754. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.L.; Nogueira, C.; Heffernan, T.P.; van Doorn, R.; Dhakal, S.; Hanna, J.A.; Min, C.; Jaskelioff, M.; Xiao, Y.; Wu, C.J.; et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell 2011, 20, 92–103. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Taylor, M.D.; Singh, P.K.; Yang, S. How does fascin promote cancer metastasis? FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zou, J.; Wang, G. Advances in the proteomic discovery of novel therapeutic targets in cancer. Drug Des. Dev. 2013, 7, 1259–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa-Zamora, P.; García-Solano, J.; García-García, F.; Turpin, M.e.C.; Trujillo-Santos, J.; Torres-Moreno, D.; Oviedo-Ramírez, I.; Carbonell-Muñoz, R.; Muñoz-Delgado, E.; Rodriguez-Braun, E.; et al. Expression profiling shows differential molecular pathways and provides potential new diagnostic biomarkers for colorectal serrated adenocarcinoma. Int. J. Cancer 2013, 132, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Alburquerque-González, B.; Bernabé-García, M.; Montoro-García, S.; Bernabé-García, Á.; Rodrigues, P.C.; Ruiz Sanz, J.; López-Calderón, F.F.; Luque, I.; Nicolas, F.J.; Cayuela, M.L.; et al. New role of the antidepressant imipramine as a Fascin1 inhibitor in colorectal cancer cells. Exp. Mol. Med. 2020, 52, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montoro-García, S.; Alburquerque-González, B.; Bernabé-García, Á.; Bernabé-García, M.; Rodrigues, P.C.; den-Haan, H.; Luque, I.; Nicolás, F.J.; Pérez-Sánchez, H.; Cayuela, M.L.; et al. Novel anti-invasive properties of a Fascin1 inhibitor on colorectal cancer cells. J. Mol. Med. 2020, 98, 383–394. [Google Scholar] [CrossRef]
- Chen, L.; Yang, S.; Jakoncic, J.; Zhang, J.J.; Huang, X.Y. Migrastatin analogues target fascin to block tumour metastasis. Nature 2010, 464, 1062–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Dalvit, C.; Fogliatto, G.; Stewart, A.; Veronesi, M.; Stockman, B. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef]
- Jansen, S.; Collins, A.; Yang, C.; Rebowski, G.; Svitkina, T.; Dominguez, R. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 2011, 286, 30087–30096. [Google Scholar] [CrossRef] [Green Version]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Jelassi, B.; Chantôme, A.; Alcaraz-Pérez, F.; Baroja-Mazo, A.; Cayuela, M.L.; Pelegrin, P.; Surprenant, A.; Roger, S. P2X(7) receptor activation enhances SK3 channels- and cystein cathepsin-dependent cancer cells invasiveness. Oncogene 2011, 30, 2108–2122. [Google Scholar] [CrossRef] [Green Version]
- Fior, R.; Póvoa, V.; Mendes, R.V.; Carvalho, T.; Gomes, A.; Figueiredo, N.; Ferreira, M.G. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl. Acad. Sci. USA 2017, 114, E8234–E8243. [Google Scholar] [CrossRef] [Green Version]
- Albini, A. Tumor and endothelial cell invasion of basement membranes. The matrigel chemoinvasion assay as a tool for dissecting molecular mechanisms. Pathol. Oncol. Res. 1998, 4, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Sliwinska, P.; Scapozza, L.; Ruiz i Altaba, A. Drug repurposing in oncology: Compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 434–454. [Google Scholar] [CrossRef] [PubMed]
- Carleo, M.A.; Di Martino, F.; Del Giudice, A.; Gargiulo, M.; Parrella, G.; Rosario, P.; Sangiovanni, V.; Viglietti, R.; Esposito, V.; Chirianni, A. Different impact of anti-retroviral regimen containing protease inhibitors on development of HIV-related Kaposi sarcoma. Vivo 2015, 29, 133–136. [Google Scholar]
- Singh, I.R.; Gorzynski, J.E.; Drobysheva, D.; Bassit, L.; Schinazi, R.F. Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome. PLoS ONE 2010, 5, e9948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.C.; Chiou, J.; Yang, Y.F.; Su, C.Y.; Lin, Y.F.; Yang, C.N.; Lu, P.J.; Huang, M.S.; Yang, C.J.; Hsiao, M. Therapeutic Targeting of Aldolase A Interactions Inhibits Lung Cancer Metastasis and Prolongs Survival. Cancer Res. 2019, 79, 4754–4766. [Google Scholar] [CrossRef] [Green Version]
- Scanlon, K.J.; Kashani-Sabet, M.; Sowers, L.C. Overexpression of DNA replication and repair enzymes in cisplatin-resistant human colon carcinoma HCT8 cells and circumvention by azidothymidine. Cancer Commun. 1989, 1, 269–275. [Google Scholar]
- Oprea, T.I.; Bauman, J.E.; Bologa, C.G.; Buranda, T.; Chigaev, A.; Edwards, B.S.; Jarvik, J.W.; Gresham, H.D.; Haynes, M.K.; Hjelle, B.; et al. Drug Repurposing from an Academic Perspective. Drug. Discov. Today Strateg. 2011, 8, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lu, Y.; Zhang, C.; Huang, D.; Wu, W.; Shen, J.; Cai, Y.; Chen, W.; Yao, W. FSCN-1 increases doxorubicin resistance in hepatocellular carcinoma through promotion of epithelial-mesenchymal transition. Int. J. Oncol. 2018, 52, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Ghebeh, H.; Al-Khaldi, S.; Olabi, S.; Al-Dhfyan, A.; Al-Mohanna, F.; Barnawi, R.; Tulbah, A.; Al-Tweigeri, T.; Ajarim, D.; Al-Alwan, M. Fascin is involved in the chemotherapeutic resistance of breast cancer cells predominantly via the PI3K/Akt pathway. Br. J. Cancer 2014, 111, 1552–1561. [Google Scholar] [CrossRef] [Green Version]
- Mohr, C.F.; Gross, C.; Bros, M.; Reske-Kunz, A.B.; Biesinger, B.; Thoma-Kress, A.K. Regulation of the tumor marker Fascin by the viral oncoprotein Tax of human T-cell leukemia virus type 1 (HTLV-1) depends on promoter activation and on a promoter-independent mechanism. Virology 2015, 485, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.L.; Miao, H.; Xia, J.; Jia, F.; Wang, H.; Xu, F.; Guo, L. Proteomics Analysis Identifies IRSp53 and Fascin as Critical for PRV Egress and Direct Cell-Cell Transmission. Proteomics 2019, 19, e1900009. [Google Scholar] [CrossRef]
- Mohr, C.F.; Kalmer, M.; Gross, C.; Mann, M.C.; Sterz, K.R.; Kieser, A.; Fleckenstein, B.; Kress, A.K. The tumor marker Fascin is induced by the Epstein-Barr virus-encoded oncoprotein LMP1 via NF-κB in lymphocytes and contributes to their invasive migration. Cell Commun. Signal. 2014, 12, 46. [Google Scholar] [CrossRef] [Green Version]
- Mosialos, G.; Yamashiro, S.; Baughman, R.W.; Matsudaira, P.; Vara, L.; Matsumura, F.; Kieff, E.; Birkenbach, M. Epstein-Barr virus infection induces expression in B lymphocytes of a novel gene encoding an evolutionarily conserved 55-kilodalton actin-bundling protein. J. Virol. 1994, 68, 7320–7328. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.P.; Chen, J.N.; Xiao, L.; He, Q.; Feng, Z.Y.; Zhang, Z.G.; Liu, J.P.; Wei, H.B.; Shao, C.K. The implication of tumor-infiltrating lymphocytes in Epstein-Barr virus-associated gastric carcinoma. Hum. Pathol. 2019, 85, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Al-Antary, N.; Farghaly, H.; Aboulkassim, T.; Yasmeen, A.; Akil, N.; Al Moustafa, A.E. Epstein-Barr virus and its association with Fascin expression in colorectal cancers in the Syrian population: A tissue microarray study. Hum. Vaccines Immunother. 2017, 13, 1573–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Said, J.W.; Pinkus, J.L.; Yamashita, J.; Mishalani, S.; Matsumura, F.; Yamashiro, S.; Pinkus, G.S. The role of follicular and interdigitating dendritic cells in HIV-related lymphoid hyperplasia: Localization of fascin. Mod. Pathol. 1997, 10, 421–427. [Google Scholar]
- Xiao, P.; Liu, W.; Zhou, H. miR-200b inhibits migration and invasion in non-small cell lung cancer cells via targeting FSCN1. Mol. Med. Rep. 2016, 14, 1835–1840. [Google Scholar] [CrossRef]
- Yang, X.; Lei, P.; Huang, Y.; Zhang, Z.; Zhang, Y. MicroRNA-133b inhibits the migration and invasion of non small cell lung cancer cells via targeting FSCN1. Oncol. Lett. 2016, 12, 3619–3625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, X.; Wu, H.; Zhou, W.; Liu, Z. MicroRNA-145 inhibits migration and invasion via inhibition of fascin 1 protein expression in non-small-cell lung cancer cells. Mol. Med. Rep. 2015, 12, 6193–6198. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, T. Downregulation of MicroRNA-135 Promotes Sensitivity of Non-Small Cell Lung Cancer to Gefitinib by Targeting TRIM16. Oncol. Res. 2018, 26, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Huang, C.; Gunda, V.; Sun, J.; Chellappan, S.P.; Li, Z.; Izumi, V.; Fang, B.; Koomen, J.; Singh, P.K.; et al. Fascin Controls Metastatic Colonization and Mitochondrial Oxidative Phosphorylation by Remodeling Mitochondrial Actin Filaments. Cell Rep. 2019, 28, 2824–2836.e2828. [Google Scholar] [CrossRef] [PubMed]








| Control | 30 µM RAL | 10 ng/mL EGF | 50 µM MEKi | 100 µM MGS | |
|---|---|---|---|---|---|
| Lamellipodium number | 8.7 ± 1.73 | 6.73 ± 1.11 | 11.58± 2.35 | 4.14 ± 1.43 | 4.58 ± 1.39 |
| p value * | 3.73 × 10−5 | 0.000026 | 5.76 × 10−10 | 3.14 ×10−9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alburquerque-González, B.; Bernabé-García, Á.; Bernabé-García, M.; Ruiz-Sanz, J.; López-Calderón, F.F.; Gonnelli, L.; Banci, L.; Peña-García, J.; Luque, I.; Nicolás, F.J.; et al. The FDA-Approved Antiviral Raltegravir Inhibits Fascin1-Dependent Invasion of Colorectal Tumor Cells In Vitro and In Vivo. Cancers 2021, 13, 861. https://doi.org/10.3390/cancers13040861
Alburquerque-González B, Bernabé-García Á, Bernabé-García M, Ruiz-Sanz J, López-Calderón FF, Gonnelli L, Banci L, Peña-García J, Luque I, Nicolás FJ, et al. The FDA-Approved Antiviral Raltegravir Inhibits Fascin1-Dependent Invasion of Colorectal Tumor Cells In Vitro and In Vivo. Cancers. 2021; 13(4):861. https://doi.org/10.3390/cancers13040861
Chicago/Turabian StyleAlburquerque-González, Begoña, Ángel Bernabé-García, Manuel Bernabé-García, Javier Ruiz-Sanz, Fernando Feliciano López-Calderón, Leonardo Gonnelli, Lucia Banci, Jorge Peña-García, Irene Luque, Francisco José Nicolás, and et al. 2021. "The FDA-Approved Antiviral Raltegravir Inhibits Fascin1-Dependent Invasion of Colorectal Tumor Cells In Vitro and In Vivo" Cancers 13, no. 4: 861. https://doi.org/10.3390/cancers13040861
APA StyleAlburquerque-González, B., Bernabé-García, Á., Bernabé-García, M., Ruiz-Sanz, J., López-Calderón, F. F., Gonnelli, L., Banci, L., Peña-García, J., Luque, I., Nicolás, F. J., Cayuela-Fuentes, M. L., Luchinat, E., Pérez-Sánchez, H., Montoro-García, S., & Conesa-Zamora, P. (2021). The FDA-Approved Antiviral Raltegravir Inhibits Fascin1-Dependent Invasion of Colorectal Tumor Cells In Vitro and In Vivo. Cancers, 13(4), 861. https://doi.org/10.3390/cancers13040861