Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness
Abstract
:Simple Summary
Abstract
1. Introduction
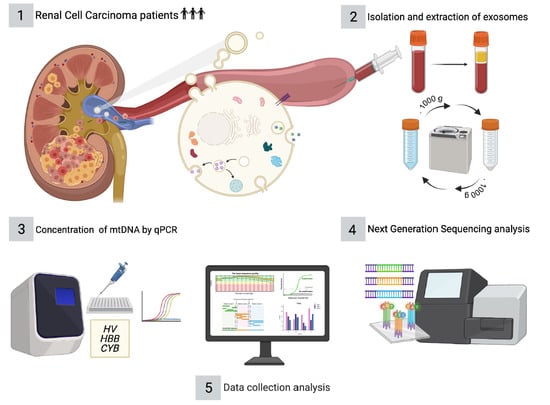
2. Materials and Methods
2.1. Patient Selection and Sample Collection
2.2. Isolation and Extraction of Exosomes from Plasma Samples
2.3. Characterization of Phase F by NanoSight LM10-HS
2.4. Absolute Quantification of mtDNA in a Control Sample by Digital PCR (dPCR)
2.5. Determination of the Relative Concentration of mtDNA by Real Time PCR (qPCR)
2.6. Next Generation Sequencing (NGS) Analyses and Data Processing
2.7. Statistical Analysis
3. Results
3.1. NTA Analysis
3.2. mtDNA in a Control Sample by dPCR
3.3. mtDNA in Relation to Aggressiveness
3.4. NGS Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef]
- Meyer, A.R.; Allaf, M.E.; Gorin, M.A. Epidemiology and Risk Factors of Renal Cell Carcinoma. In Diagnosis and Surgical Management of Renal Tumors; Gorin, M.A., Allaf, M.E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–11. [Google Scholar]
- Oto, J.; Plana, E.; Vicente Sanchez-Gonzalez, J.; Garcia-Olaverri, J.; Fernandez-Pardo, A.; Espana, F.; Martinez-Sarmiento, M.; Vera-Donoso, C.D.; Navarro, S.; Medina, P. Urinary microRNAs: Looking for a New Tool in Diagnosis, Prognosis, and Monitoring of Renal Cancer. Curr. Urol. Rep. 2020, 21, 11. [Google Scholar] [CrossRef]
- Signoretti, S.; Flaifel, A.; Chen, Y.; Reuter, V.E. Renal Cell Carcinoma in the Era of Precision Medicine: From Molecular Pathology to Tissue-Based Biomarkers. J. Clin. Oncol. 2018, 36, 3553. [Google Scholar] [CrossRef] [PubMed]
- Dudani, S.; Savard, M.; Heng, D.Y.C. An Update on Predictive Biomarkers in Metastatic Renal Cell Carcinoma. Eur. Urol. Focus 2020, 6, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Farber, N.J.; Kim, C.J.; Modi, P.K.; Hon, J.D.; Sadimin, E.T.; Singer, E.A. Renal Cell Carcinoma: The Search for a Reliable Biomarker. Transl. Cancer Res. 2017, 6, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Shan, H.; Miao, M.Z.; Jiang, Z.; Meng, Y.; Chen, R.; Zhang, L.; Liu, Y. Identification of the Key Genes and Pathways Involved in the Tumorigenesis and Prognosis of Kidney Renal Clear Cell Carcinoma. Sci. Rep. 2020, 10, 4271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, L.; Wu, P.; Li, S.; Wei, D. Overexpression of MAX Dimerization Protein 3 (MXD3) Predicts Poor Prognosis in Clear Cell Renal Cell Carcinoma. Transl. Androl. Urol. 2021, 10, 785–796. [Google Scholar] [CrossRef]
- Shu, X.; Gu, J.; Huang, M.; Tannir, N.M.; Matin, S.F.; Karam, J.A.; Wood, C.G.; Wu, X.; Ye, Y. Germline Genetic Variants in Somatically Significantly Mutated Genes in Tumors Are Associated with Renal Cell Carcinoma Risk and Outcome. Carcinogenesis 2018, 39, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Pido, S.; Ceddia, G.; Masseroli, M. Computational Analysis of Fused Co-Expression Networks for the Identification of Candidate Cancer Gene Biomarkers. NPJ Syst. Biol. Appl. 2021, 7, 17. [Google Scholar] [CrossRef]
- Lin, Y.; Yang, Z.; Xu, A.; Dong, P.; Huang, Y.; Liu, H.; Li, F.; Wang, H.; Xu, Q.; Wang, Y.; et al. PIK3R1 Negatively Regulates the Epithelial-Mesenchymal Transition and Stem-Like Phenotype of Renal Cancer Cells through the AKT/GSK3β/CTNNB1 Signaling Pathway. Sci. Rep. 2015, 5, 8997. [Google Scholar] [CrossRef] [PubMed]
- Mytsyk, Y.; Borys, Y.; Tumanovska, L.; Stroy, D.; Kucher, A.; Gazdikova, K.; Rodrigo, L.; Kruzliak, P.; Prosecky, R.; Urdzik, P.; et al. MicroRNA-15a Tissue Expression Is a Prognostic Marker for Survival in Patients with Clear Cell Renal Cell Carcinoma. Clin. Exp. Med. 2019, 19, 515–524. [Google Scholar] [CrossRef]
- Tang, T.; Du, X.; Zhang, X.; Niu, W.; Li, C.; Tan, J. Computational Identification and Analysis of Early Diagnostic Biomarkers for Kidney Cancer. J. Hum. Genet. 2019, 64, 1015–1022. [Google Scholar] [CrossRef]
- Connor, K.L.; Denby, L. MicroRNAs as Non-Invasive Biomarkers of Renal Disease. Nephrol. Dial. Transplant. 2021, 36, 428–429. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, Y.; Zhao, E.; Tan, Y.; Geng, B.; Kang, C.; Li, X. PTRF/CAVIN1, Regulated by SHC1 through the EGFR Pathway, Is Found in Urine Exosomes as a Potential Biomarker of ccRCC. Carcinogenesis 2020, 41, 274–283. [Google Scholar] [CrossRef]
- Fujitaka, K.; Murakami, T.; Takeuchi, M.; Kakimoto, T.; Mochida, H.; Arakawa, K. mRNAs in Urinary Nano-Extracellular Vesicles as Potential Biomarkers for Non-Invasive Kidney Biopsy. Biomed. Rep. 2021, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Long, M.; Yu, G.; Cheng, Y.; Yang, Q.; Liu, J.; Wang, Y.; Sheng, J.; Wang, L.; Wang, Z.; et al. Urinary Exosome miR-30c-5p as a Biomarker of Clear Cell Renal Cell Carcinoma that Inhibits Progression by Targeting HSPA5. J. Cell. Mol. Med. 2019, 23, 6755–6765. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nishiyama, A.; Matsuyama, M.; Wang, Z.; Yuan, Y. The (Pro)Renin Receptor: A Novel Biomarker and Potential Therapeutic Target for Various Cancers. Cell Commun. Signal. 2020, 18, 39. [Google Scholar] [CrossRef] [Green Version]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Pan, M.; Chen, P.; Lu, J.; Liu, Z.; Jia, E.; Ge, Q. The Fragmentation Patterns of Maternal Plasma Cell-Free DNA and Its Applications in Non-Invasive Prenatal Testing. Prenat. Diagn. 2020, 40, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Tsui, N.B.Y.; Jiang, P.; Chow, K.C.K.; Su, X.; Leung, T.Y.; Sun, H.; Chan, K.C.A.; Chiu, R.W.K.; Lo, Y.M.D. High Resolution Size Analysis of Fetal DNA in the Urine of Pregnant Women by Paired-End Massively Parallel Sequencing. PLoS ONE 2012, 7, e48319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valpione, S.; Gremel, G.; Mundra, P.; Middlehurst, P.; Galvani, E.; Girotti, M.R.; Lee, R.J.; Garner, G.; Dhomen, N.; Lorigan, P.C.; et al. Plasma Total Cell-Free DNA (cfDNA) Is a Surrogate Biomarker for Tumour Burden and a Prognostic Biomarker for Survival in Metastatic Melanoma Patients. Eur. J. Cancer 2018, 88, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yu, M. Circulating Cell-Free Mitochondrial DNA as a Novel Cancer Biomarker: Opportunities and Challenges. Mitochondrial DNA 2012, 23, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Barekati, Z.; Radpour, R.; Zhong, X.Y. Cell-Free DNA in the Circulation as a Potential Cancer Biomarker. Anticancer Res. 2011, 31, 2623–2628. [Google Scholar]
- Yang, K.; Li, X.; Forman, M.R.; Monahan, P.O.; Graham, B.H.; Joshi, A.; Song, M.; Hang, D.; Ogino, S.; Giovannucci, E.L.; et al. Pre-Diagnostic Leukocyte Mitochondrial DNA Copy Number and Colorectal Cancer Risk. Carcinogenesis 2019, 40, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Platel, M.; Mahasheh, A.; Ambrosone, C.B.; Zhao, H. Mitochondrial Copy Number and Risk of Breast Cancer: A Pilot Study. Mitochondrion 2010, 10, 62–68. [Google Scholar] [CrossRef] [Green Version]
- Gentiluomo, M.; Katzke, V.A.; Kaaks, R.; Tjonneland, A.; Severi, G.; Perduca, V.; Boutron-Ruault, M.; Weiderpass, E.; Ferrari, P.; Johnson, T.; et al. Mitochondrial DNA Copy-Number Variation and Pancreatic Cancer Risk in the Prospective EPIC Cohort. Cancer Epidemiol. Biomark. Prev. 2020, 29, 681–686. [Google Scholar] [CrossRef]
- Oliveira, G.L.; Coelho, A.R.; Marques, R.; Oliveira, P.J. Cancer Cell Metabolism: Rewiring the Mitochondrial Hub. Biochim. Biophys. Acta-Mol. Basis Dis. 2021, 1867, 166016. [Google Scholar] [CrossRef]
- Freeman, B.; Smith, N.; Curtis, C.; Huckett, L.; Mill, J.; Craig, I.W. DNA from Buccal Swabs Recruited by Mail: Evaluation of Storage Effects on Long-Term Stability and Suitability for Multiplex Polymerase Chain Reaction Genotyping. Behav. Genet. 2003, 33, 67–72. [Google Scholar] [CrossRef]
- Gomez-Martin, A.; Hernandez, A.F.; Javier Martinez-Gonzalez, L.; Gonzalez-Alzaga, B.; Rodriguez-Barranco, M.; Lopez-Flores, I.; Aguilar-Garduno, C.; Lacasana, M. Polymorphisms of Pesticide-Metabolizing Genes in Children Living in Intensive Farming Communities. Chemosphere 2015, 139, 534–540. [Google Scholar] [CrossRef]
- ThermoFisher Scientific. QuantStudio™ 3D Digital PCR System User Guide. 2015. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0007720.pdf (accessed on 22 June 2021).
- Bio-Rad. iTaq Universal Probes One-Step Kit, Vol. Mix, No. Table 1. Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/10032046.pdf (accessed on 22 June 2021).
- Carvalho, A.; Ferreira, G.; Seixas, D.; Guimaraes-Teixeira, C.; Henrique, R.; Monteiro, F.J.; Jeronimo, C. Emerging Lab-on-a-Chip Approaches for Liquid Biopsy in Lung Cancer: Status in CTCs and ctDNA Research and Clinical Validation. Cancers 2021, 13, 2101. [Google Scholar] [CrossRef]
- Ciferri, M.C.; Quarto, R.; Tasso, R. Extracellular Vesicles as Biomarkers and Therapeutic Tools: From Pre-Clinical to Clinical Applications. Biology 2021, 10, 359. [Google Scholar] [CrossRef]
- Pozo-Agundo, A.; Villaescusa, N.; Martorell-Marugán, J.; Soriano, O.; Leyva, S.; Jódar-Reyes, A.B.; Botella, L.M.; Carmona-Sáez, P.; Blanco, F.J. Identification of Exosomal MicroRNA Signature by Liquid Biopsy in Hereditary Hemorrhagic Telangiectasia Patients. Int. J. Mol. Sci. 2021, 22, 9450. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, A.; Solimando, A.G.; Kerscher, A.; Bittrich, M.; Kalogirou, C.; Kubler, H.; Rosenwald, A.; Bargou, R.; Kollmannsberger, P.; Schilling, B.; et al. Subgroup-Independent Mapping of Renal Cell Carcinoma-Machine Learning Reveals Prognostic Mitochondrial Gene Signature Beyond Histopathologic Boundaries. Front. Oncol. 2021, 11, 621278. [Google Scholar] [CrossRef] [PubMed]
- DiNatale, R.G.; Sanchez, A.; Hakimi, A.A.; Reznik, E. Metabolomics Informs Common Patterns of Molecular Dysfunction Across Histologies of Renal Cell Carcinoma. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 755–762. [Google Scholar] [CrossRef]
- Bond, K.H.; Fetting, J.L.; Lary, C.W.; Emery, I.F.; Oxburgh, L. FOXD1 Regulates Cell Division in Clear Cell Renal Cell Carcinoma. BMC Cancer 2021, 21, 312. [Google Scholar] [CrossRef]
- Xu, H.; Zou, R.; Li, F.; Liu, J.; Luan, N.; Wang, S.; Zhu, L. MRPL15 Is a Novel Prognostic Biomarker and Therapeutic Target for Epithelial Ovarian Cancer. Cancer Med. 2021, 10, 3655–3673. [Google Scholar] [CrossRef]
- Zhao, Z.; He, K.; Zhang, Y.; Hua, X.; Feng, M.; Zhao, Z.; Sun, Y.; Jiang, Y.; Xia, Q. XRCC2 Repairs Mitochondrial DNA Damage and Fuels Malignant Behavior in Hepatocellular Carcinoma. Cancer Lett. 2021, 512, 1–14. [Google Scholar] [CrossRef]
- Valencia, K.; Montuenga, L.M. Exosomes in Liquid Biopsy: The Nanometric World in the Pursuit of Precision Oncology. Cancers 2021, 13, 2147. [Google Scholar] [CrossRef] [PubMed]
- Lazar, S.; Goldfinger, L.E. Platelets and Extracellular Vesicles and Their Cross Talk with Cancer. Blood 2021, 137, 3192–3200. [Google Scholar] [CrossRef]
- Muramatsu-Maekawa, Y.; Kawakami, K.; Fujita, Y.; Takai, M.; Kato, D.; Nakane, K.; Kato, T.; Tsuchiya, T.; Koie, T.; Miura, Y.; et al. Profiling of Serum Extracellular Vesicles Reveals miRNA-4525 as a Potential Biomarker for Advanced Renal Cell Carcinoma. Cancer Genom. Proteom. 2021, 18, 253–259. [Google Scholar] [CrossRef]
- Song, W.; Chen, Y.; Zhu, G.; Xie, H.; Yang, Z.; Li, L. Exosome-Mediated miR-9-5p Promotes Proliferation and Migration of Renal Cancer Cells both In Vitro and In Vivo by Targeting SOCS4. Biochem. Biophys. Res. Commun. 2020, 529, 1216–1224. [Google Scholar] [CrossRef]
- Dias, F.; Teixeira, A.L.; Nogueira, I.; Morais, M.; Maia, J.; Bodo, C.; Ferreira, M.; Silva, A.; Vilhena, M.; Lobo, J.; et al. Extracellular Vesicles Enriched in Hsa-miR-301a-3p and Hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers 2020, 12, 1450. [Google Scholar] [CrossRef] [PubMed]
- Dias, F.; Teixeira, A.L.; Nogueira, I.; Morais, M.; Maia, J.; Bodo, C.; Ferreira, M.; Vieira, I.; Silva, J.; Lobo, J.; et al. Plasma Extracellular Vesicle-Derived TIMP-1 mRNA as a Prognostic Biomarker in Clear Cell Renal Cell Carcinoma: A Pilot Study. Int. J. Mol. Sci. 2020, 21, 4624. [Google Scholar] [CrossRef] [PubMed]
- VanEtten, S.L.; Bonner, M.R.; Ren, X.; Birnbaum, L.S.; Kostyniak, P.J.; Wang, J.; Olson, J.R. Effect of Exposure to 2,3,7,8-Tetrachlorodibenzo-P-Dioxin (TCDD) and Polychlorinated Biphenyls (PCBs) on Mitochondrial DNA (mtDNA) Copy Number in Rats. Toxicology 2021, 454, 152744. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Ganguly, S.; Palanichamy, J.K.; Singh, A.; Bakhshi, R.; Jain, A.; Chopra, A.; Bakhshi, S. PGC1A Driven Enhanced Mitochondrial DNA Copy Number Predicts Outcome in Pediatric Acute Myeloid Leukemia. Mitochondrion 2021, 58, 246–254. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, K.; Guo, S.; Wang, Y.; Ji, X.; Yuan, Q.; Su, L.; Guo, X.; Gu, X.; Xing, J. NGS-Based Accurate and Efficient Detection of Circulating Cell-Free Mitochondrial DNA in Cancer Patients. Mol. Ther.-Nucleic Acids 2021, 23, 657–666. [Google Scholar] [CrossRef]
- Rai, N.K.; Panjwani, G.; Ghosh, A.K.; Haque, R.; Sharma, L.K. Analysis of Mitochondrial DNA Copy Number Variation in Blood and Tissue Samples of Metastatic Breast Cancer Patients (A Pilot Study). Biochem. Biophys. Rep. 2021, 26, 100931. [Google Scholar] [CrossRef]
- Dache, Z.A.A.; Otandault, A.; Tanos, R.; Pastor, B.; Meddeb, R.; Sanchez, C.; Arena, G.; Lasorsa, L.; Bennett, A.; Grange, T.; et al. Blood Contains Circulating Cell-Free Respiratory Competent Mitochondria. FASEB J. 2020, 34, 3616–3630. [Google Scholar] [CrossRef] [Green Version]
- Elsayed, E.T.; Hashad, M.M.; Elgohary, I.E. Mitochondrial DNA Copy Number Variation as a Potential Predictor of Renal Cell Carcinoma. Int. J. Biol. Mark. 2017, 32, E313–E318. [Google Scholar] [CrossRef]


| Characteristic | Patients (N = 13) | Controls (N = 15) |
|---|---|---|
| Age (yr) | ||
| Median (range)–yr | 68 (47–88) | 67 (44–93) |
| <65 yr | 5 | 6 |
| >65 yr | 8 | 9 |
| Sex | ||
| Male | 11 | 7 |
| Female | 2 | 8 |
| Histology | ||
| Papillary | 1 | NA |
| Clear cell | 12 | NA |
| Size tumor (cm) | 9 (5–18) | NA |
| Stage | ||
| Stage III | 7 | |
| Stage IV | 6 | |
| TNM | NA | |
| T1 | 1 | |
| T2 | 1 | |
| T3 | 10 | |
| T4 | 1 | |
| Fuhrman nuclear grade | NA | |
| G3 | 2 | |
| G4 | 11 | |
| Metastasis | NA | |
| No | 7 | |
| Yes | 6 |
| Fraction | Sample | Obtaining |
|---|---|---|
| B | 200 µL plasma | Plasma obtained after centrifugation (1400× g, 4 °C, 10 min) |
| C | Pellet | Pellet obtained after centrifugation with DTT + PBS (16,000× g, 4 °C, 20 min) |
| D | 200 µL supernatant | Supernatant obtained after centrifugation (15,000× g, 4 °C, 30 min) |
| E | 6 mL supernatant | Supernatant obtained after ultracentrifugation (160,000× g, 4 °C, 2 h) |
| F | Pellet | Pellet obtained after ultracentrifugation (160,000× g, 4 °C, 2 h) |
| Phase | Gene | Adjusted p Value (*) | Adjusted p Value (cn) | Ct Mean No Metastasis ± SD | Ct Mean Metastasis ± SD | Copies Per µL Mean No Metastasis ± SD | Copies Per µL Mean Metastasis ± SD |
|---|---|---|---|---|---|---|---|
| B | HV1-short | 0.020 | 0.069 | 24.54 ± 4.18 | 22.06 ± 2.08 | 2.38 ± 3.77 | 2.55 ± 2.04 |
| HV1-long | 0.035 | 0.133 | 23.92 ± 4.61 | 21.35 ± 2.56 | 2.67 ± 3.92 | 2.56 ± 1.72 | |
| CYB-short | 0.078 | 0.223 | 25.71 ± 4.29 | 23.69 ± 2.54 | 2.08 ± 3.11 | 1.90 ± 1.35 | |
| HBB-long | 0.020 | 0.029 | 33.52 ± 2.99 | 36.09 ± 2.69 | 1.07 ± 2.02 | 0.09 ± 0.16 | |
| C | CYB-short | 0.359 | 0.037 | 19.95 ± 2.73 | 20.67 ± 2.03 | 24.36 ± 29.66 | 10.18 ± 7.37 |
| HBB-short | 0.001 | 0.002 | 19.18 ± 2.41 | 21.24 ± 0.89 | 1092.33 ± 1254.73 | 194.36 ± 102.38 | |
| HBB-long | 0.006 | 0.012 | 30.37 ± 3.71 | 32.99 ± 1.55 | 23.39 ± 49.39 | 0.38 ± 0.48 | |
| D | HBB-long | 0.001 | 0.001 | 31.84 ± 3.48 | 36.03 ± 2.77 | 7.39 ± 15.66 | 0.07 ± 0.09 |
| F | HBB-long | 0.007 | 0.014 | 31.72 ± 2.13 | 33.44 ± 1.34 | 2.22 ± 4.42 | 0.22 ± 0.19 |
| Phase | % Mapped | Mapping Quality | Percentage in Genome | RPKM |
|---|---|---|---|---|
| B | 99 | 28.65 | Autosomes chr. 95.575% Sexual Chr. 4.387% mtDNA 0.038% | 0.320 0.173 22.382 |
| C | 89 | 28.575 | Autosomes chr. 95.331% Sexual Chr. 4.308% mtDNA 0.361% | 0.316 0.179 212.287 |
| D | 98 | 29.055 | Autosomes chr. 95.647% Sexual Chr. 4.344% mtDNA 0.010% | 0.322 0.176 5.598 |
| E | 100 | 29.65 | Autosomes chr. 95.665% Sexual Chr. 4.333% mtDNA 0.002% | 0.319 0.177 0.437 |
| F | 94 | 29.535 | Autosomes chr. 95.800% Sexual Chr. 4.184% mtDNA 0.016% | 0.325 0.167 9.303 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arance, E.; Ramírez, V.; Rubio-Roldan, A.; Ocaña-Peinado, F.M.; Romero-Cachinero, C.; Jódar-Reyes, A.B.; Vazquez-Alonso, F.; Martinez-Gonzalez, L.J.; Alvarez-Cubero, M.J. Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness. Cancers 2022, 14, 199. https://doi.org/10.3390/cancers14010199
Arance E, Ramírez V, Rubio-Roldan A, Ocaña-Peinado FM, Romero-Cachinero C, Jódar-Reyes AB, Vazquez-Alonso F, Martinez-Gonzalez LJ, Alvarez-Cubero MJ. Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness. Cancers. 2022; 14(1):199. https://doi.org/10.3390/cancers14010199
Chicago/Turabian StyleArance, Elena, Viviana Ramírez, Alejandro Rubio-Roldan, Francisco M. Ocaña-Peinado, Catalina Romero-Cachinero, Ana Belén Jódar-Reyes, Fernando Vazquez-Alonso, Luis Javier Martinez-Gonzalez, and Maria Jesus Alvarez-Cubero. 2022. "Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness" Cancers 14, no. 1: 199. https://doi.org/10.3390/cancers14010199
APA StyleArance, E., Ramírez, V., Rubio-Roldan, A., Ocaña-Peinado, F. M., Romero-Cachinero, C., Jódar-Reyes, A. B., Vazquez-Alonso, F., Martinez-Gonzalez, L. J., & Alvarez-Cubero, M. J. (2022). Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness. Cancers, 14(1), 199. https://doi.org/10.3390/cancers14010199