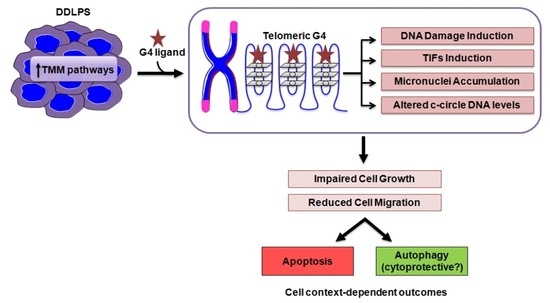
Telomere as a Therapeutic Target in Dedifferentiated Liposarcoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Chemicals
2.2. Gene Expression Profiles
2.3. Characterization of TMM in DDLPS Cells
2.4. Assessment of the Expression Levels of Telomerase Components by Real-Time RT-PCR
2.5. Cell-Based Experiments
2.6. Western Immunoblotting
2.7. Fluorescence Microscopy Analyses
2.8. Statistical Analysis
3. Results
3.1. “Telomere Maintenance” Pathways Are Up-Regulated in DDLPS
3.2. Characterization of the Operating TMM in DDLPS Cell Lines
3.3. Biological Effects of Equitoxic Concentrations of RHPS4 in DDLPS Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jo, V.Y.; Fletcher, C.D.M. WHO classification of soft tissue tumours: An update based on the 2013 (4th) edition. Pathology 2014, 46, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Amin-Mansour, A.; George, S.; Sioletic, S.; Carter, S.L.; Rosenberg, M.; Taylor-Weiner, A.; Stewart, C.; Chevalier, A.; Seepo, S.; Tracy, A.; et al. Genomic Evolutionary Patterns of Leiomyosarcoma and Liposarcoma. Clin. Cancer Res. 2019, 25, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Thway, K. Well-differentiated liposarcoma and dedifferentiated liposarcoma: An updated review. Semin. Diagn. Pathol. 2019, 36, 112–121. [Google Scholar] [CrossRef]
- Hirata, M.; Asano, N.; Katayama, K.; Yoshida, A.; Tsuda, Y.; Sekimizu, M.; Mitani, S.; Kobayashi, E.; Komiyama, M.; Fujimoto, H.; et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat. Commun. 2019, 10, 5683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronchi, A.; Strauss, D.C.; Miceli, R.; Bonvalot, S.; Swallow, C.J.; Hohenberger, P.; Van Coevorden, F.; Rutkowski, P.; Callegaro, D.; Hayes, A.J.; et al. Variability in Patterns of Recurrence After Resection of Primary Retroperitoneal Sarcoma (RPS): A Report on 1007 Patients From the Multi-institutional Collaborative RPS Working Group. Ann. Surg. 2016, 263, 1002–1009. [Google Scholar] [CrossRef]
- Van Houdt, W.J.; Zaidi, S.; Messiou, C.; Thway, K.; Strauss, D.C.; Jones, R.L. Treatment of retroperitoneal sarcoma: Current standards and new developments. Curr. Opin. Oncol. 2017, 29, 260–267. [Google Scholar] [CrossRef]
- Italiano, A.; Toulmonde, M.; Cioffi, A.; Penel, N.; Isambert, N.; Bompas, E.; Duffaud, F.; Patrikidou, A.; Lortal, B.; Le Cesne, A.; et al. Advanced well-differentiated/dedifferentiated liposarcomas: Role of chemotherapy and survival. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 1601–1607. [Google Scholar] [CrossRef]
- Jones, R.L.; Fisher, C.; Al-Muderis, O.; Judson, I.R. Differential sensitivity of liposarcoma subtypes to chemotherapy. Eur. J. Cancer 2005, 41, 2853–2860. [Google Scholar] [CrossRef]
- Tap, W.D.; Wagner, A.J.; Schöffski, P.; Martin-Broto, J.; Krarup-Hansen, A.; Ganjoo, K.N.; Yen, C.C.; Abdul Razak, A.R.; Spira, A.; Kawai, A.; et al. Effect of Doxorubicin Plus Olaratumab vs Doxorubicin Plus Placebo on Survival in Patients With Advanced Soft Tissue Sarcomas: The ANNOUNCE Randomized Clinical Trial. JAMA 2020, 323, 1266–1276. [Google Scholar] [CrossRef]
- Gahvari, Z.; Parkes, A. Dedifferentiated Liposarcoma: Systemic Therapy Options. Curr. Treat. Options Oncol. 2020, 21, 15. [Google Scholar] [CrossRef]
- Callegaro, D.; Raut, C.P.; Ng, D.; Strauss, D.C.; Honoré, C.; Stoeckle, E.; Bonvalot, S.; Haas, R.L.; Vassos, N.; Conti, L.; et al. Has the Outcome for Patients Who Undergo Resection of Primary Retroperitoneal Sarcoma Changed Over Time? A Study of Time Trends During the Past 15 years. Ann. Surg. Oncol. 2021, 28, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Recagni, M.; Bidzinska, J.; Zaffaroni, N.; Folini, M. The role of alternative lengthening of telomeres mechanism in cancer: Translational and therapeutic implications. Cancers 2020, 12, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, J.D.; Reddel, R.R. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010, 584, 3800–3811. [Google Scholar] [CrossRef] [Green Version]
- Sommer, A.; Royle, N.J. ALT: A multi-faceted phenomenon. Genes 2020, 11, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, J.D.; Cao, Y.; Huschtscha, L.I.; Chang, A.C.; Au, A.Y.M.; Pickett, H.A.; Reddel, R.R. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009, 27, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Dagg, R.; Zhang, Y.; Lee, J.H.Y.; Lu, R.; Martin La Rotta, N.; Sampl, S.; Korkut-Demirba, M.; Holzmann, K.; Lau, L.M.S.; et al. The C-Circle Biomarker Is Secreted by Alternative-Lengthening-of-Telomeres Positive Cancer Cells inside Exosomes and Provides a Blood-Based Diagnostic for ALT Activity. Cancers 2021, 13, 5369. [Google Scholar] [CrossRef]
- Robinson, N.J.; Schiemann, W.P. Means to the ends: The role of telomeres and telomere processing machinery in metastasis. Biochim. Biophys. Acta-Rev. Cancer 2016, 1866, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Teber, E.T.; Holmes, O.; Nones, K.; Patch, A.M.; Dagg, R.A.; Lau, L.M.S.; Lee, J.H.; Napier, C.E.; Arthur, J.W.; et al. Telomere sequence content can be used to determine ALT activity in tumours. Nucleic Acids Res. 2018, 46, 4903–4918. [Google Scholar] [CrossRef] [Green Version]
- Guterres, A.N.; Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Relitti, N.; Saraswati, A.P.; Federico, S.; Khan, T.; Brindisi, M.; Zisterer, D.; Brogi, S.; Gemma, S.; Butini, S.; Campiani, G. Telomerase-based Cancer Therapeutics: A Review on their Clinical Trials. Curr. Top. Med. Chem. 2020, 20, 433–457. [Google Scholar] [CrossRef]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, I.; Recagni, M.; Zaffaroni, N.; Folini, M. On the Road to Fight Cancer: The Potential of G-Quadruplex Ligands as Novel Therapeutic Agents. Int. J. Mol. Sci. 2021, 22, 5947. [Google Scholar] [CrossRef] [PubMed]
- Liau, J.-Y.; Lee, J.-C.; Tsai, J.-H.; Yang, C.-Y.; Liu, T.-L.; Ke, Z.-L.; Hsu, H.-H.; Jeng, Y.-M. Comprehensive screening of alternative lengthening of telomeres phenotype and loss of ATRX expression in sarcomas. Mod. Pathol. 2015, 28, 1545–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.E.; Varkonyi, R.J.; Schwalm, J.; Cragle, R.; Klein-Szanto, A.; Patchefsky, A.; Cukierman, E.; Von Mehren, M.; Broccoli, D. Multiple Mechanisms of Telomere Maintenance Exist in Liposarcomas. Clin. Cancer Res. 2005, 11, 5347–5355. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Martin, V.; Soriano, M.; Garcia-Salcedo, J.A. Quadruplex ligands in cancer therapy. Cancers 2021, 13, 3156. [Google Scholar] [CrossRef]
- Cookson, J.C.; Dai, F.; Smith, V.; Heald, R.A.; Laughton, C.A.; Stevens, M.F.G.; Burger, A.M. Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: Activity in human tumor cells correlates with telomere length and can be enhanced, or antagoni. Mol. Pharmacol. 2005, 68, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Zuco, V.; Pasquali, S.; Tortoreto, M.; Brich, S.; Percio, S.; Dagrada, G.P.; Colombo, C.; Sanfilippo, R.; Lauricella, C.; Gounder, M.; et al. Selinexor versus doxorubicin in dedifferentiated liposarcoma PDXs: Evidence of greater activity and apoptotic response dependent on p53 nuclear accumulation and survivin down-regulation. J. Exp. Clin. Cancer Res. 2021, 40, 83. [Google Scholar] [CrossRef]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Cimino-Reale, G.; Gandellini, P.; Santambrogio, F.; Recagni, M.; Zaffaroni, N.; Folini, M. MiR-380-5p-mediated repression of TEP1 and TSPYL5 interferes with telomerase activity and favours the emergence of an “ALT-like” phenotype in diffuse malignant peritoneal mesothelioma cells. J. Hematol. Oncol. 2017, 10, 140. [Google Scholar] [CrossRef] [Green Version]
- Henson, J.D.; Lau, L.M.; Koch, S.; Martin La Rotta, N.; Dagg, R.A.; Reddel, R.R. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 2017, 114, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Roake, C.M.; Freund, A.; Batista, P.J.; Tian, S.; Yin, Y.A.; Gajera, C.R.; Lin, S.; Lee, B.; Pech, M.F.; et al. An Activity Switch in Human Telomerase Based on RNA Conformation and Shaped by TCAB1. Cell 2018, 174, 218–230.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saharia, A.; Stewart, S.A. FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene 2009, 28, 1162–1167. [Google Scholar] [CrossRef] [Green Version]
- Sampathi, S.; Chai, W. Mapping the FEN1 interaction domain with hTERT. Biochem. Biophys. Res. Commun. 2011, 407, 34–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivashkevich, A.; Redon, C.E.; Nakamura, A.J.; Martin, R.F.; Martin, O.A. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012, 327, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of Telomeres pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef]
- Rizzo, A.; Salvati, E.; Biroccio, A. Methods of studying telomere damage induced by quadruplex-ligand complexes. Methods 2012, 57, 93–99. [Google Scholar] [CrossRef]
- Barth, S.; Glick, D.; Macleod, K.F. Autophagy: Assays and artifacts. J. Pathol. 2010, 221, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Eskelinen, E.L.; Tanaka, Y.; Saftig, P. At the acidic edge: Emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003, 13, 137–145. [Google Scholar] [CrossRef]
- Lewis, C.W.; Golsteyn, R.M. Cancer cells that survive checkpoint adaptation contain micronuclei that harbor damaged DNA. Cell Cycle 2016, 15, 3131–3145. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; Harrington, L.; Baird, D.M. Chromothripsis and telomere crisis: Engines of genome instability. Curr. Opin. Genet. Dev. 2020, 60, 41–47. [Google Scholar] [CrossRef] [PubMed]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [Green Version]
- Miglietta, G.; Russo, M.; Duardo, R.C.; Capranico, G. G-quadruplex binders as cytostatic modulators of innate immune genes in cancer cells. Nucleic Acids Res. 2021, 49, 6673–6686. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Miglietta, G.; Morigi, R.; Iaccarino, N.; Locatelli, A.; Leoni, A.; Novellino, E.; Pagano, B.; Capranico, G.; Randazzo, A. Monohydrazone Based G-Quadruplex Selective Ligands Induce DNA Damage and Genome Instability in Human Cancer Cells. J. Med. Chem. 2020, 63, 3090–3103. [Google Scholar] [CrossRef]
- Asaithamby, A.; Hu, B.; Chen, D.J. Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8293–8298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conomos, D.; Pickett, H.A.; Reddel, R.R.; Azzalin, C. Alternative lengthening of telomeres: Remodeling the telomere architecture. Front. Oncol. 2013, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Salimi-Jeda, A.; Ghabeshi, S.; Gol Mohammad pour, Z.; Jazaeri, E.O.; Araiinejad, M.; Sheikholeslami, F.; Abdoli, M.; Edalat, M.; Abdoli, A. Autophagy Modulation and Cancer Combination Therapy: A Smart Approach in Cancer Therapy. Cancer Treat. Res. Commun. 2022, 30, 100512. [Google Scholar] [CrossRef]
- Amato, R.; Valenzuela, M.; Berardinelli, F.; Salvati, E.; Maresca, C.; Leone, S.; Antoccia, A.; Sgura, A. G-quadruplex stabilization fuels the ALT pathway in ALT-positive osteosarcoma cells. Genes 2020, 11, 304. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Ni, J.; Liang, Z.; Xue, J.; Fenech, M.F.; Wang, X. The molecular origins and pathophysiological consequences of micronuclei: New insights into an age-old problem. Mutat. Res.-Rev. Mutat. Res. 2019, 779, 1–35. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; D’Angelo, S.; Attia, S.; Riedel, R.F.; Priebat, D.A.; et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): A multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet. Oncol. 2017, 18, 1493–1501. [Google Scholar] [CrossRef]
- Voorwerk, L.; Slagter, M.; Horlings, H.M.; Sikorska, K.; van de Vijver, K.K.; de Maaker, M.; Nederlof, I.; Kluin, R.J.C.; Warren, S.; Ong, S.F.; et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: The TONIC trial. Nat. Med. 2019, 25, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.J.; Ajani, J.A.; Kuzdzal, J.; Zander, T.; Van Cutsem, E.; Piessen, G.; Mendez, G.; Feliciano, J.; Motoyama, S.; Lièvre, A.; et al. Adjuvant Nivolumab in Resected Esophageal or Gastroesophageal Junction Cancer. N. Engl. J. Med. 2021, 384, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.K.; Zybailov, B.L.; Maddukuri, L.; Gao, J.; Marecki, J.C.; Jaiswal, M.; Bell, M.R.; Griffin, W.C.; Reed, M.R.; Chib, S.; et al. Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress. J. Biol. Chem. 2016, 291, 18041–18057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.H.; Nie, X.; Liu, H.Y.; Fang, Y.M.; Zhao, Y.; Xia, L.X. TMPyP4 promotes cancer cell migration at low doses, but induces cell death at high doses. Sci. Rep. 2016, 6, 26592. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Cell Line | 48 h | 72 h |
|---|---|---|
| LS-GD-1 | 3.2 ± 0.3 | 1.7 ± 0.1 |
| LS-BZ-1 | 4.1 ± 0.2 | 2.5 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alessandrini, I.; Percio, S.; Naghshineh, E.; Zuco, V.; Stacchiotti, S.; Gronchi, A.; Pasquali, S.; Zaffaroni, N.; Folini, M. Telomere as a Therapeutic Target in Dedifferentiated Liposarcoma. Cancers 2022, 14, 2624. https://doi.org/10.3390/cancers14112624
Alessandrini I, Percio S, Naghshineh E, Zuco V, Stacchiotti S, Gronchi A, Pasquali S, Zaffaroni N, Folini M. Telomere as a Therapeutic Target in Dedifferentiated Liposarcoma. Cancers. 2022; 14(11):2624. https://doi.org/10.3390/cancers14112624
Chicago/Turabian StyleAlessandrini, Irene, Stefano Percio, Eisa Naghshineh, Valentina Zuco, Silvia Stacchiotti, Alessandro Gronchi, Sandro Pasquali, Nadia Zaffaroni, and Marco Folini. 2022. "Telomere as a Therapeutic Target in Dedifferentiated Liposarcoma" Cancers 14, no. 11: 2624. https://doi.org/10.3390/cancers14112624
APA StyleAlessandrini, I., Percio, S., Naghshineh, E., Zuco, V., Stacchiotti, S., Gronchi, A., Pasquali, S., Zaffaroni, N., & Folini, M. (2022). Telomere as a Therapeutic Target in Dedifferentiated Liposarcoma. Cancers, 14(11), 2624. https://doi.org/10.3390/cancers14112624