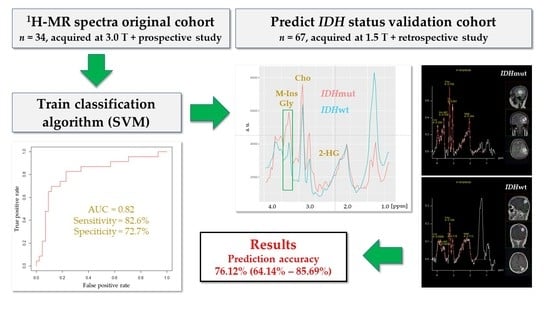
Validation Study for Non-Invasive Prediction of IDH Mutation Status in Patients with Glioma Using In Vivo 1H-Magnetic Resonance Spectroscopy and Machine Learning
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Validation Cohort
2.2. Neuropathological Assessment of Glioma Tissue
2.2.1. Immunohistochemistry
2.2.2. DNA Extraction and Quantification
2.2.3. Next-Generation Sequencing (NGS)
2.2.4. Multiplex Ligation-Dependent Probe Amplification (MLPA)
2.3. Data Acquisition of 1H-MRS
2.4. H-MRS Data Preprocessing
2.5. H-MRS Classification
3. Results
3.1. Patient Characteristics of the Validation Cohort
3.2. IDH Mutation Caused Specific Alterations in 1H-MR Spectra
3.3. A Linear SVM Provided High Sensitivity and Specificity in Detecting IDHmut
3.4. SVM Did Not Predict LOH 1p/19q Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro-Oncology 2020, 22, 1073–1113. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs. Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Yuan, Y.; Wu, J.; Mendoza, T.; Vera, E.; Omuro, A.; Lieberman, F.; Robins, H.I.; Gerstner, E.R.; Wu, J.; et al. A phase II study of dose-dense temozolomide and lapatinib for recurrent low-grade and anaplastic supratentorial, infratentorial, and spinal cord ependymoma. Neuro-Oncology 2021, 23, 468–477. [Google Scholar] [CrossRef]
- Cairncross, G.; Wang, M.; Shaw, E.; Jenkins, R.; Brachman, D.; Buckner, J.; Fink, K.; Souhami, L.; Laperriere, N.; Curran, W.; et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: Long-term results of RTOG 9402. J. Clin. Oncol. 2013, 31, 337–343. [Google Scholar] [CrossRef]
- Van den Bent, M.J.; Brandes, A.A.; Taphoorn, M.J.; Kros, J.M.; Kouwenhoven, M.C.; Delattre, J.Y.; Bernsen, H.J.; Frenay, M.; Tijssen, C.C.; Grisold, W.; et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: Long-term follow-up of EORTC brain tumor group study 26951. J. Clin. Oncol. 2013, 31, 344–350. [Google Scholar] [CrossRef]
- Platten, M.; Bunse, L.; Wick, A.; Bunse, T.; Le Cornet, L.; Harting, I.; Sahm, F.; Sanghvi, K.; Tan, C.L.; Poschke, I.; et al. A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature 2021, 592, 463–468. [Google Scholar] [CrossRef]
- Kaminska, B.; Czapski, B.; Guzik, R.; Król, S.K.; Gielniewski, B. Consequences of IDH1/2 Mutations in Gliomas and an Assessment of Inhibitors Targeting Mutated IDH Proteins. Molecules 2019, 24, 968. [Google Scholar] [CrossRef] [Green Version]
- Ellingson, B.M.; Wen, P.Y.; Cloughesy, T.F. Modified Criteria for Radiographic Response Assessment in Glioblastoma Clinical Trials. Neurotherapeutics 2017, 14, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Pasquini, L.; Napolitano, A.; Tagliente, E.; Dellepiane, F.; Lucignani, M.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Moltoni, G.; Nicolai, M.; et al. Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. J. Pers. Med. 2021, 11, 290. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Lucignani, M.; Tagliente, E.; Dellepiane, F.; Rossi-Espagnet, M.C.; Ritrovato, M.; Vidiri, A.; Villani, V.; Ranazzi, G.; et al. AI and High-Grade Glioma for Diagnosis and Outcome Prediction: Do All Machine Learning Models Perform Equally Well? Front. Oncol. 2021, 11, 601425. [Google Scholar] [CrossRef]
- Sotoudeh, H.; Shafaat, O.; Bernstock, J.D.; Brooks, M.D.; Elsayed, G.A.; Chen, J.A.; Szerip, P.; Chagoya, G.; Gessler, F.; Sotoudeh, E.; et al. Artificial Intelligence in the Management of Glioma: Era of Personalized Medicine. Front. Oncol. 2019, 9, 768. [Google Scholar] [CrossRef]
- Suh, C.H.; Kim, H.S.; Jung, S.C.; Choi, C.G.; Kim, S.J. 2-Hydroxyglutarate MR spectroscopy for prediction of isocitrate dehydrogenase mutant glioma: A systemic review and meta-analysis using individual patient data. Neuro-Oncology 2018, 20, 1573–1583. [Google Scholar] [CrossRef] [Green Version]
- Berrington, A.; Voets, N.L.; Larkin, S.J.; de Pennington, N.; McCullagh, J.; Stacey, R.; Schofield, C.J.; Jezzard, P.; Clare, S.; Cadoux-Hudson, T.; et al. A comparison of 2-hydroxyglutarate detection at 3 and 7 T with long-TE semi-LASER. NMR Biomed. 2018, 31, e3886. [Google Scholar] [CrossRef]
- Shen, X.; Voets, N.L.; Larkin, S.J.; de Pennington, N.; Plaha, P.; Stacey, R.; McCullagh, J.S.O.; Schofield, C.J.; Clare, S.; Jezzard, P.; et al. A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy. Metabolites 2019, 9, 35. [Google Scholar] [CrossRef] [Green Version]
- Askari, P.; Dimitrov, I.E.; Ganji, S.K.; Tiwari, V.; Levy, M.; Patel, T.R.; Pan, E.; Mickey, B.E.; Malloy, C.R.; Maher, E.A.; et al. Spectral fitting strategy to overcome the overlap between 2-hydroxyglutarate and lipid resonances at 2.25 ppm. Magn. Reson. Med. 2021, 86, 1818–1828. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Kim, G.S.; Gerstner, E.; Batchelor, T.; Tzika, A.A.; Fantin, V.R.; Vander Heiden, M.G.; Sorensen, A.G. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 2012, 4, 116ra114. [Google Scholar] [CrossRef] [Green Version]
- Verma, G.; Mohan, S.; Nasrallah, M.P.; Brem, S.; Lee, J.Y.; Chawla, S.; Wang, S.; Nagarajan, R.; Thomas, M.A.; Poptani, H. Non-invasive detection of 2-hydroxyglutarate in IDH-mutated gliomas using two-dimensional localized correlation spectroscopy (2D L-COSY) at 7 Tesla. J. Transl. Med. 2016, 14, 274. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, H.; Tanaka, K.; Sasayama, T.; Irino, Y.; Sato, N.; Takeuchi, Y.; Kyotani, K.; Mukasa, A.; Mizukawa, K.; Sakata, J.; et al. Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro-Oncology 2016, 18, 1559–1568. [Google Scholar] [CrossRef] [Green Version]
- Branzoli, F.; Pontoizeau, C.; Tchara, L.; Di Stefano, A.L.; Kamoun, A.; Deelchand, D.K.; Valabrègue, R.; Lehéricy, S.; Sanson, M.; Ottolenghi, C.; et al. Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro-Oncology 2019, 21, 765–774. [Google Scholar] [CrossRef]
- Smits, M. Imaging of oligodendroglioma. Br. J. Radiol. 2016, 89, 20150857. [Google Scholar] [CrossRef]
- Bumes, E.; Wirtz, F.P.; Fellner, C.; Grosse, J.; Hellwig, D.; Oefner, P.J.; Häckl, M.; Linker, R.; Proescholdt, M.; Schmidt, N.O.; et al. Non-Invasive Prediction of IDH Mutation in Patients with Glioma WHO II/III/IV Based on F-18-FET PET-Guided In Vivo (1)H-Magnetic Resonance Spectroscopy and Machine Learning. Cancers 2020, 12, 3406. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Wen, P.Y.; Huse, J.T. 2016 World Health Organization Classification of Central Nervous System Tumors. Continuum 2017, 23, 1531–1547. [Google Scholar] [CrossRef]





| IDH Mutation | ||||
|---|---|---|---|---|
| IDHwt | IDHmut | |||
| Number | In % | Number | In % | |
| Sex | ||||
| men | 26 | 59% | 15 | 65% |
| women | 18 | 41% | 8 | 35% |
| WHO | ||||
| II | 3 | 7% | 13 | 57% |
| III | 4 | 9% | 9 | 39% |
| IV | 36 | 82% | 1 | 4% |
| unknown | 1 | 2% | 0 | 0% |
| LOH1p19q | ||||
| no | 4 | 9% | 10 | 43% |
| yes | 0 | 0% | 7 | 30% |
| unknown | 40 | 91% | 6 | 26% |
| Total | 44 | 23 | ||
| Original Cohort | Validation Cohort | |
|---|---|---|
| Value (95% CI) | Value (95% CI) | |
| Accuracy (%) | 88.24 (72.55–96.70) | 76.12 (64.14–85.69) |
| Sensitivity (%) | 95.45 (77.16–99.88) | 82.61 (61.22–95.05) |
| Specificity (%) | 75.00 (42.81 94.51) | 72.73 (57.21–85.04) |
| Positive likelihood ratio | 3.82 (1.43–10.22) | 3.03 (1.80–5.08) |
| Negative likelihood ratio | 0.06 (0.01–0.42) | 0.24 (0.10–0.59) |
| Positive predictive value (%) | 87.50 (72.35–94.93) | 61.29 (48.55–72.66) |
| Negative predictive value (%) | 90.00 (56.33–98.43) | 88.89 (76.32–95.20) |
| Disease prevalence (%) | 64.71 (46.49–80.25) | 34.33 (23.15–46.94) |
| AUC | 0.83 | 0.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bumes, E.; Fellner, C.; Fellner, F.A.; Fleischanderl, K.; Häckl, M.; Lenz, S.; Linker, R.; Mirus, T.; Oefner, P.J.; Paar, C.; et al. Validation Study for Non-Invasive Prediction of IDH Mutation Status in Patients with Glioma Using In Vivo 1H-Magnetic Resonance Spectroscopy and Machine Learning. Cancers 2022, 14, 2762. https://doi.org/10.3390/cancers14112762
Bumes E, Fellner C, Fellner FA, Fleischanderl K, Häckl M, Lenz S, Linker R, Mirus T, Oefner PJ, Paar C, et al. Validation Study for Non-Invasive Prediction of IDH Mutation Status in Patients with Glioma Using In Vivo 1H-Magnetic Resonance Spectroscopy and Machine Learning. Cancers. 2022; 14(11):2762. https://doi.org/10.3390/cancers14112762
Chicago/Turabian StyleBumes, Elisabeth, Claudia Fellner, Franz A. Fellner, Karin Fleischanderl, Martina Häckl, Stefan Lenz, Ralf Linker, Tim Mirus, Peter J. Oefner, Christian Paar, and et al. 2022. "Validation Study for Non-Invasive Prediction of IDH Mutation Status in Patients with Glioma Using In Vivo 1H-Magnetic Resonance Spectroscopy and Machine Learning" Cancers 14, no. 11: 2762. https://doi.org/10.3390/cancers14112762
APA StyleBumes, E., Fellner, C., Fellner, F. A., Fleischanderl, K., Häckl, M., Lenz, S., Linker, R., Mirus, T., Oefner, P. J., Paar, C., Proescholdt, M. A., Riemenschneider, M. J., Rosengarth, K., Weis, S., Wendl, C., Wimmer, S., Hau, P., Gronwald, W., & Hutterer, M. (2022). Validation Study for Non-Invasive Prediction of IDH Mutation Status in Patients with Glioma Using In Vivo 1H-Magnetic Resonance Spectroscopy and Machine Learning. Cancers, 14(11), 2762. https://doi.org/10.3390/cancers14112762