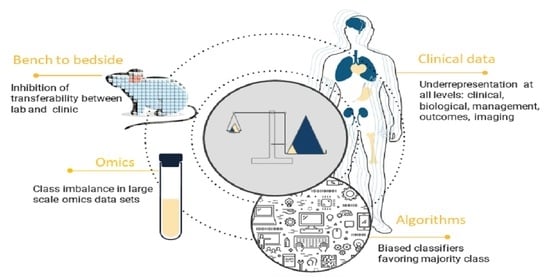
Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Sources of Bias and Class Imbalance in Large Scale Medical Data
2.1. Gender and Race Imbalance
2.2. Limited Capture and Inclusion of Social Determinants of Health (SDOH) and Disability
2.3. Variability in Oncologic Management
2.4. Variability in Oncologic Outcomes Capture
2.5. Knowledge Translation Bias
3. Overview of the Existing Methods for the Class Imbalance Problem
3.1. Data-Level Methods
3.2. Algorithm-Level Methods
3.2.1. Recognition-Based Methods
3.2.2. Cost-Sensitive Methods
3.2.3. Ensemble Learning-Based Methods
3.3. Hybrid Methods
3.4. Other Methods
4. Guidelines to Mitigate Bias and Address Class Imbalance in Oncologic Data Sets towards Transferrable AI
4.1. Clinical Data
- (1)
- Curate data with a view towards aggregation with parallel data sources that include the possibility to cross-check data collection against other data parameters, e.g., state or government data sources that can act as data verifiers of SDOH, disability, geographical location, and access to care.
- (2)
- Collect binary data when collecting more detailed data is not possible or impractical to obtain.
- (3)
- Collect data electronically with limited manual input and, when possible, automatically limit dependence on clinical provider input to allow for consistent long-term data collection as alternatives augment bias when some centers/settings collect data and others less so or not at all.
- (4)
- Implement data curation/clean data and data “health checks” for known sources of bias and class imbalance.
- (5)
- Implement capture and analysis of SDOH, disability, sex differences, ethnicity, and race data whenever possible and subject analysis to bias check, which can then be accounted for in future research and interpretation of data and resulting algorithms.
4.2. Other Data
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACC | Accuracy |
| AGC | Advanced Gastric Cancer |
| AI | Artificial Intelligence |
| AMP | Antimicrobial Peptides |
| AUPRC | Area Under Precision–Recall Curve |
| CADx | Computer-Aided Diagnosis |
| CNN | Convolutional Neural Networks |
| GBM | Glioblastoma Multiforme |
| GMEAN | Geometric Mean |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LSTM | Long Short-Term Memory |
| LVQ | Learning Vector Quantization |
| MCC | Matthews Correlation Coefficient |
| MLP | Multi-Layer Perceptron |
| MRI | Magnetic Resonance Imaging |
| NB | Naive Bayes |
| QA | Quality Assurance |
| PCA | Principal Component Analysis |
| PRE | Precision |
| PTM | Post-Translation Modification |
| REC | Recall |
| RT | Radiation Therapy |
| RUSBoost | Random Under-Sampling Boost |
| SDOH | Social Determinants of Health |
| SMOTE | Synthetic Minority Oversampling Technique |
| SOKL | Space-Occpying Kidney Lesion |
| SVM | Support Vector Machine |
| VCU | Virginia Commonwealth University |
| VHA | Veterans Health Affairs |
References
- Belenguer, L. AI bias: Exploring discriminatory algorithmic decision-making models and the application of possible machine-centric solutions adapted from the pharmaceutical industry. AI Ethic 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.K. Rising to the challenge of bias in health care AI. Nat. Med. 2021, 27, 2079–2081. [Google Scholar] [CrossRef] [PubMed]
- Daneshjou, R.; Smith, M.P.; Sun, M.D.; Rotemberg, V.; Zou, J. Lack of Transparency and Potential Bias in Artificial Intelligence Data Sets and Algorithms. JAMA Dermatol. 2021, 157, 1362. [Google Scholar] [CrossRef] [PubMed]
- Noor, P. Can we trust AI not to further embed racial bias and prejudice? BMJ 2020, 368, m363. [Google Scholar] [CrossRef] [PubMed]
- Megahed, F.M.; Chen, Y.-J.; Megahed, A.; Ong, Y.; Altman, N.; Krzywinski, M. The class imbalance problem. Nat. Methods 2021, 18, 1270–1272. [Google Scholar] [CrossRef]
- Flynn, E.; Chang, A.; Altman, R.B. Large-scale labeling and assessment of sex bias in publicly available expression data. BMC Bioinform. 2021, 22, 168. [Google Scholar] [CrossRef]
- Williamson, C.W.; Nelson, T.; Thompson, C.A.; Vitzthum, L.K.; Zakeri, K.; Riviere, P.; Bryant, A.K.; Sharabi, A.B.; Zou, J.; Mell, L.K. Bias Reduction through Analysis of Competing Events (BRACE) Correction to Address Cancer Treatment Selection Bias in Observational Data. Clin. Cancer Res. 2022, 28, 1832–1840. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 1 December 2021).
- Landers, R.N.; Behrend, T.S. Auditing the AI auditors: A framework for evaluating fairness and bias in high stakes AI predictive models. Am. Psychol. 2022. [Google Scholar] [CrossRef]
- Abd Elrahman, S.M.; Abraham, A. A Review of Class Imbalance Problem. J. Netw. Innov. Comput. 2013, 1, 332–340. [Google Scholar]
- Nguyen, G.H.; Bouzerdoum, A.; Phung, S.L. Learning Pattern Classification Tasks with Imbalanced Data Sets. In Pattern Recognition; Yin, P.-Y., Ed.; IntechOpen: Vienna, Austria, 2009. [Google Scholar] [CrossRef]
- Rajaraman, S.; Ganesan, P.; Antani, S. Deep learning model calibration for improving performance in class-imbalanced medical image classification tasks. PLoS ONE 2022, 17, e0262838. [Google Scholar] [CrossRef]
- Larrazabal, A.J.; Nieto, N.; Peterson, V.; Milone, D.H.; Ferrante, E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. 2020, 117, 12592–12594. [Google Scholar] [CrossRef] [PubMed]
- Ntoutsi, E.; Fafalios, P.; Gadiraju, U.; Iosifidis, V.; Nejdl, W.; Vidal, M.; Ruggieri, S.; Turini, F.; Papadopoulos, S.; Krasanakis, E.; et al. Bias in data-driven artificial intelligence systems—An introductory survey. WIREs Data Min. Knowl. Discov. 2020, 10, e1356. [Google Scholar] [CrossRef]
- Khushi, M.; Shaukat, K.; Alam, T.M.; Hameed, I.A.; Uddin, S.; Luo, S.; Yang, X.; Reyes, M.C. A Comparative Performance Analysis of Data Resampling Methods on Imbalance Medical Data. IEEE Access 2021, 9, 109960–109975. [Google Scholar] [CrossRef]
- Marcu, L.G. Gender and Sex-Related Differences in Normal Tissue Effects Induced by Platinum Compounds. Pharmaceuticals 2022, 15, 255. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, B.; Oertelt-Prigione, S.; Adjei, A.; Borchmann, S.; Haanen, J.; Letsch, A.; Mir, O.; Quaas, A.; Verhoeven, R.; Wagner, A. Investigation of sex and gender differences in oncology gains momentum: ESMO announces the launch of a Gender Medicine Task Force. Ann. Oncol. 2022, 33, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, A.C.; Gozzi, B.; Cardoso-Filho, C.; Machado, H.; Zeferino, L.C.; Vale, D.B. Race disparities in mortality by breast cancer from 2000 to 2017 in São Paulo, Brazil: A population-based retrospective study. BMC Cancer 2021, 21, 998. [Google Scholar] [CrossRef]
- Morshed, R.A.; Reihl, S.J.; Molinaro, A.M.; Kakaizada, S.; Young, J.S.; Schulte, J.D.; Butowski, N.; Taylor, J.; Bush, N.A.; Aghi, M.K.; et al. The influence of race and socioeconomic status on therapeutic clinical trial screening and enrollment. J. Neuro-Oncol. 2020, 148, 131–139. [Google Scholar] [CrossRef]
- Ragavan, M.; Patel, M.I. The evolving landscape of sex-based differences in lung cancer: A distinct disease in women. Eur. Respir. Rev. 2022, 31, 210100. [Google Scholar] [CrossRef]
- Mazul, A.L.; Naik, A.N.; Zhan, K.Y.; Stepan, K.O.; Old, M.O.; Kang, S.Y.; Nakken, E.R.; Puram, S.V. Gender and race interact to influence survival disparities in head and neck cancer. Oral Oncol. 2021, 112, 105093. [Google Scholar] [CrossRef]
- Carrano, A.; Juarez, J.; Incontri, D.; Ibarra, A.; Cazares, H.G. Sex-Specific Differences in Glioblastoma. Cells 2021, 10, 1783. [Google Scholar] [CrossRef]
- Massey, S.C.; Whitmire, P.; Doyle, T.E.; Ippolito, J.E.; Mrugala, M.M.; Hu, L.S.; Canoll, P.; Anderson, A.R.; Wilson, M.A.; Fitzpatrick, S.M.; et al. Sex differences in health and disease: A review of biological sex differences relevant to cancer with a spotlight on glioma. Cancer Lett. 2020, 498, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Tucker-Seeley, R.D. Social Determinants of Health and Disparities in Cancer Care for Black People in the United States. JCO Oncol. Pract. 2021, 17, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Leech, M.M.; Weiss, J.E.; Markey, C.; Loehrer, A.P. Influence of Race, Insurance, Rurality, and Socioeconomic Status on Equity of Lung and Colorectal Cancer Care. Ann. Surg. Oncol. 2022, 29, 3630–3639. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Natwick, T.; Liu, J.; Morrison, V.A.; Vidito, S.; Werther, W.; Yusuf, A.A.; Usmani, S.Z. Mortality by a proxy performance status as defined by a claims-based measure for disability status in older patients with newly diagnosed multiple myeloma in the United States. J. Geriatr. Oncol. 2019, 10, 490–496. [Google Scholar] [CrossRef]
- Al Feghali, K.A.; Buszek, S.M.; Elhalawani, H.; Chevli, N.; Allen, P.K.; Chung, C. Real-world evaluation of the impact of radiotherapy and chemotherapy in elderly patients with glioblastoma based on age and performance status. Neuro-Oncol. Pract. 2020, 8, 199–208. [Google Scholar] [CrossRef]
- Jung, H.; Lu, M.; Quan, M.L.; Cheung, W.Y.; Kong, S.; Lupichuk, S.; Feng, Y.; Xu, Y. New method for determining breast cancer recurrence-free survival using routinely collected real-world health data. BMC Cancer 2022, 22, 281. [Google Scholar] [CrossRef]
- Rossi, L.A.; Melstrom, L.G.; Fong, Y.; Sun, V. Predicting post-discharge cancer surgery complications via telemonitoring of patient-reported outcomes and patient-generated health data. J. Surg. Oncol. 2021, 123, 1345–1352. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Höhn, J.; Hekler, A.; Krieghoff-Henning, E.; Kather, J.N.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hauschild, A.; French, L.; Schlager, J.G.; et al. Skin Cancer Classification Using Convolutional Neural Networks with Integrated Patient Data: A Systematic Review (Preprint). J. Med Internet Res. 2020, 23, e20708. [Google Scholar] [CrossRef]
- Wong, A.; Otles, E.; Donnelly, J.P.; Krumm, A.; McCullough, J.; DeTroyer-Cooley, O.; Pestrue, J.; Phillips, M.; Konye, J.; Penoza, C.; et al. External Validation of a Widely Implemented Proprietary Sepsis Prediction Model in Hospitalized Patients. JAMA Intern. Med. 2021, 181, 1065–1070. [Google Scholar] [CrossRef]
- Vyas, D.A.; Eisenstein, L.G.; Jones, D.S. Hidden in Plain Sight—Reconsidering the Use of Race Correction in Clinical Algorithms. New Engl. J. Med. 2020, 383, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Microsoft; Abernethy, A.; Shaywitz, D.; Gundlapalli, A.; Weinstein, J.; Doraiswamy, P.M.; Schulman, K.; Madhavan, S.; Verily; et al. Digital Health COVID-19 Impact Assessment: Lessons Learned and Compelling Needs. NAM Perspect. 2022. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.S.W.; Syed, K.; Hagan, M.; Palta, J.; Kapoor, R.; Ghosh, P. Deep neural network models to automate incident triage in the radiation oncology incident learning system. In Proceedings of the BCB’21: Proceedings of the 12th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, Gainesville, FL, USA, 1–4 August 2021; Volume 51, pp. 1–10. [Google Scholar]
- Brown, W.E.; Sung, K.; Aleman, D.M.; Moreno-Centeno, E.; Purdie, T.; McIntosh, C.J. Guided undersampling classification for automated radiation therapy quality assurance of prostate cancer treatment. Med Phys. 2018, 45, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hall, L.O.; Bowyer, K.W.; Goldgof, D.B.; Gatenby, R.; Ben Ahmed, K. Synthetic minority image over-sampling technique: How to improve AUC for glioblastoma patient survival prediction. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 1357–1362. [Google Scholar] [CrossRef]
- Suárez-García, J.G.; Hernández-López, J.M.; Moreno-Barbosa, E.; de Celis-Alonso, B. A simple model for glioma grading based on texture analysis applied to conventional brain MRI. PLoS ONE 2020, 15, e0228972. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tao, Y.; Cong, H.; Zhu, E.; Cai, T. Predicting liver cancers using skewed epidemiological data. Artif. Intell. Med. 2022, 124, 102234. [Google Scholar] [CrossRef]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; pp. 287–297. [Google Scholar] [CrossRef]
- Goyal, S.; Raghunathan, A.; Jain, M.; Simhadri, H.V.; Jain, P. DROCC: Deep Robust One-Class Classification. In Proceedings of the 37th International Conference on Machine Learning, Proceedings of Machine Learning Research, Online, 13–18 July 2020; pp. 3711–3721. [Google Scholar]
- Gao, L.; Zhang, L.; Liu, C.; Wu, S. Handling imbalanced medical image data: A deep-learning-based one-class classification approach. Artif. Intell. Med. 2020, 108, 101935. [Google Scholar] [CrossRef]
- Welch, M.L.; McIntosh, C.; McNiven, A.; Huang, S.H.; Zhang, B.-B.; Wee, L.; Traverso, A.; O’Sullivan, B.; Hoebers, F.; Dekker, A.; et al. User-controlled pipelines for feature integration and head and neck radiation therapy outcome predictions. Phys. Medica 2020, 70, 145–152. [Google Scholar] [CrossRef]
- Leevy, J.; Khoshgoftaar, T.M.; Bauder, R.A.; Seliya, N. A survey on addressing high-class imbalance in big data. J. Big Data 2018, 5, 42. [Google Scholar] [CrossRef]
- Nguyen, N.N.; Duong, A.T. Comparison of Two Main Approaches for Handling Imbalanced Data in Churn Prediction Problem. J. Adv. Inf. Technol. 2021, 12. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.B.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar]
- Jaeger, P.F.; Kohl, S.A.A.; Bickelhaupt, S.; Isensee, F.; Kuder, T.A.; Schlemmer, H.-P.; Maier-Hein, K.H. Retina U-Net: Embarrassingly Simple Exploitation of Segmentation Supervision for Medical Object Detection. In Proceedings of the Machine Learning for Health NeurIPS Workshop, Proceedings of Machine Learning Research; 2020; pp. 171–183. Available online: http://proceedings.mlr.press/v116/jaeger20a/jaeger20a.pdf (accessed on 1 December 2021).
- Xiong, Y.; Ye, M.; Wu, C. Cancer Classification with a Cost-Sensitive Naive Bayes Stacking Ensemble. Comput. Math. Methods Med. 2021, 2021, 5556992. [Google Scholar] [CrossRef]
- Shon, H.S.; Batbaatar, E.; Kim, K.O.; Cha, E.J.; Kim, K.-A. Classification of Kidney Cancer Data Using Cost-Sensitive Hybrid Deep Learning Approach. Symmetry 2020, 12, 154. [Google Scholar] [CrossRef]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2019, 14, 241–258. [Google Scholar] [CrossRef]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Tang, X.; Cai, L.; Meng, Y.; Gu, C.; Yang, J.; Yang, J. A Novel Hybrid Feature Selection and Ensemble Learning Framework for Unbalanced Cancer Data Diagnosis With Transcriptome and Functional Proteomic. IEEE Access 2021, 9, 51659–51668. [Google Scholar] [CrossRef]
- Le, D.N.T.; Le, H.X.; Ngo, L.; Ngo, H.T. Transfer learning with class-weighted and focal loss function for automatic skin cancer classification. arXiv 2020, arXiv:2009.05977. [Google Scholar]
- Wang, S.; Dong, D.; Zhang, W.; Hu, H.; Li, H.; Zhu, Y.; Zhou, J.; Shan, X.; Tian, J. Specific Borrmann classification in advanced gastric cancer by an ensemble multilayer perceptron network: A multicenter research. Med Phys. 2021, 48, 5017–5028. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. Using Random Forest to Learn Imbalanced Data. Univ. Calif. Berkeley 2004, 110, 24. [Google Scholar]
- Zhao, D.; Liu, H.; Zheng, Y.; He, Y.; Lu, D.; Lyu, C. Whale optimized mixed kernel function of support vector machine for colorectal cancer diagnosis. J. Biomed. Informatics 2019, 92, 103124. [Google Scholar] [CrossRef]
- Urdal, J.; Engan, K.; Kvikstad, V.; Janssen, E.A. Prognostic prediction of histopathological images by local binary patterns and RUSBoost. In Proceedings of the 2017 25th European Signal Processing Conference (EUSIPCO), Kos, Greece, 28 August–2 September 2017. [Google Scholar] [CrossRef]
- Mirza, B.; Wang, W.; Wang, J.; Choi, H.; Chung, N.C.; Ping, P. Machine Learning and Integrative Analysis of Biomedical Big Data. Genes 2019, 10, 87. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Hilario, M.; Kalousis, A. Approaches to dimensionality reduction in proteomic biomarker studies. Briefings Bioinform. 2007, 9, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B-Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Yan, K.K.; Zhao, H.; Pang, H. A comparison of graph- and kernel-based–omics data integration algorithms for classifying complex traits. BMC Bioinform. 2017, 18, 539. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An Introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Garcıa, V.; Sánchez, S.J.; Mollineda, R.A. Exploring the Performance of Resampling Strategies for the Class Imbalance Problem; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Lao, J.; Chen, Y.; Li, Z.-C.; Li, Q.; Zhang, J.; Liu, J.; Zhai, G. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci. Rep. 2017, 7, 10353. [Google Scholar] [CrossRef]
- Wu, A.; Li, Y.; Qi, M.; Lu, X.; Jia, Q.; Guo, F.; Dai, Z.; Liu, Y.; Chen, C.; Zhou, L.; et al. Dosiomics improves prediction of locoregional recurrence for intensity modulated radiotherapy treated head and neck cancer cases. Oral Oncol. 2020, 104, 104625. [Google Scholar] [CrossRef]




| Study | Technique | Setting |
|---|---|---|
| Data-level methods | ||
| Bose et al. [35] | Convolutional neural networks (CNN), long short-term memory (LSTM), over-sampling | Radiation Oncology, incident reporting |
| Brown et al. [36] | Under-sampling | Radiation Oncology, prostate cancer |
| Liu et al. [37] | Synthetic Minority Oversampling Technique (SMOTE) | Glioblastoma imaging |
| Suarez-Garcia et al. [38] | Under-sampling | Glioma imaging |
| Li et al. [39] | Under-sampling, K-Means++ and learning vector quantization (LVQ) | Liver cancer |
| Isensee et al. [40] | Deep learning, nnU-net | Brain tumor segmentation |
| Algorithm level methods | ||
| Goyal et al. [41] | Recognition-based, one class classification | real-world data sets across different domains: tabular data, images (CIFAR and ImageNet), audio, and time-series |
| Gao et al. [42] | Recognition-based, one class classification | Medical imaging |
| Welch et al. [43] | Recognition-based | Head and Neck Radiation therapy |
| Leevy et al. [44] | Cost-sensitive | General |
| Nguyen et al. [45] | Cost-sensitive, comparison of techniques | SMOTE and Deep Belief Network (DBN) against the two cost-sensitive learning methods (focal loss and weighted loss) in churn prediction problem |
| Milletari et al. [46] | Cost-sensitive | Medical image segmentation |
| Lin et al. [47] | Cost-sensitive, Focal loss | Medical imaging |
| Jaeger et al. [48] | Cost-sensitive, Focal loss | Medical imaging object detection |
| Xiong et al. [49] | Cost-Sensitive Naive Bayes Stacking Ensemble | Various malignancy data sets and data types |
| Shon et al. [50] | Cost-sensitive | Kidney cancer data (TCGA) |
| Dong et al. [51] | Ensemble-Learning | General |
| Sagi et al. [52] | Ensemble-Learning | General |
| Tang et al. [53] | Ensemble-Learning, bagging-based | Transcriptome and functional proteomics data breast cancer |
| Le et al. [54] | Ensemble-Learning, ResNet50 CNN | Skin cancer |
| Wang et al. [55] | Ensemble-Learning, multi-layer perceptron (MLP)-based | Gastric cancer |
| Hybrid methods | ||
| Khushi et al. [15] | Comparative Performance Analysis of Data Re-sampling Methods | Various malignancy data sets and data types |
| Chen et al. [56] | Combination of methods | General |
| Zhao et al. [57] | Random Under-Sampling Boost (RUSBoost) | Colorectal cancer, microarray data |
| Urdal et al. [58] | Random Under-Sampling Boost (RUSBoost) | Urothelial carcinoma, histopathology data |
| Other methods and reviews | ||
| Mirza et al. [59] | Integrative analysis of biomedical big data | |
| Guyon et al. [60] | variable and feature selection overview | |
| Hilario et al. [61] | Class imbalance in proteomic biomarker studies overview | |
| Tibshirani et al. [62] | LASSO (Least Absolute Shrinkage and Selection Operator) overview | |
| Yan et al. [63] | Graph- and kernel-based—omics data integration algorithms | |
| Fawcett et al. [64] | ROC analysis overview | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasci, E.; Zhuge, Y.; Camphausen, K.; Krauze, A.V. Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers 2022, 14, 2897. https://doi.org/10.3390/cancers14122897
Tasci E, Zhuge Y, Camphausen K, Krauze AV. Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers. 2022; 14(12):2897. https://doi.org/10.3390/cancers14122897
Chicago/Turabian StyleTasci, Erdal, Ying Zhuge, Kevin Camphausen, and Andra V. Krauze. 2022. "Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets" Cancers 14, no. 12: 2897. https://doi.org/10.3390/cancers14122897
APA StyleTasci, E., Zhuge, Y., Camphausen, K., & Krauze, A. V. (2022). Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers, 14(12), 2897. https://doi.org/10.3390/cancers14122897