Lung Adenocarcinoma Cell Sensitivity to Chemotherapies: A Spotlight on Lipid Droplets and SREBF1 Gene
Abstract
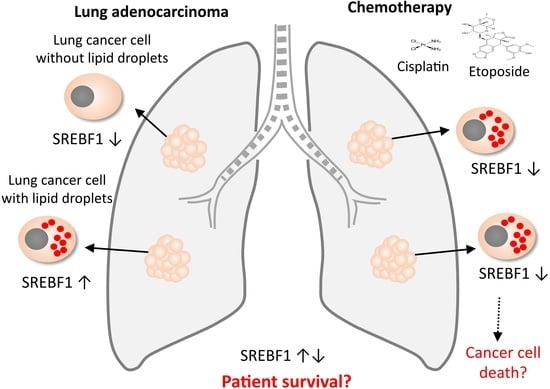
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemotherapeutic Drugs
2.2. Neutrophil Isolation and Degranulation
2.3. Cell Culture and In Vitro Experiments
2.4. RNA Isolation
2.5. Transcriptome Analysis (RNA-Seq)
2.6. RNA-Seq Data Analysis
2.7. CDNA Synthesis and Quantitative Real-Time PCR Analysis
2.8. PE Annexin V Apoptosis Detection
2.9. Metabolic Assay
2.10. Oil Red Staining
2.11. Dot Blots
2.12. Lung Cancer Patient Tissue Samples
2.13. SREBF1 Expression Analysis in Tissues
2.14. Statistical Analysis
3. Results
3.1. Missing a Clear Relationship between LD Formation, SREBF1 Expression, and Cell Sensitivity to Chemotherapeutics
3.2. Morphological Characteristics of H1299 but Not H1563 Cells Change in Serum-Free Neutrophil Degranulation Medium (NDM) as Compared to Basal Medium (BM)
3.3. The Expression of Genes Related to Lipid Metabolism Differs between H1299 and H1563 Cells
3.4. Lipid Droplets (LDs)-Bearing H1299 Cells Are More Sensitive to Etoposide Than H1563 Cells Nonbearing LDs
3.5. Cisplatin and Etoposide Induce LD Formation in H1563 Cells but Do Not Affect LD in H1299 Cells
3.6. The Lower SREBF1 Expression in Tumor Than in Nontumor Tissues from Patients with Lung Adenocarcinoma and Squamous Cell Carcinoma Correlates with a Better Overall Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Chern, Y.J.; Tai, I.T. Adaptive response of resistant cancer cells to chemotherapy. Cancer Biol. Med. 2020, 17, 842–863. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.L.S.; Barreto, E.D.A.; Fazolini, N.P.B.; Viola, J.P.B.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Petan, T. Lipid Droplets in Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–34. [Google Scholar]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef]
- Chakraborty, P.K.; Xiong, X.; Mustafi, S.B.; Saha, S.; Dhanasekaran, D.; Mandal, N.A.; McMeekin, S.; Bhattacharya, R.; Mukherjee, P. Role of cystathionine beta synthase in lipid metabolism in ovarian cancer. Oncotarget 2015, 6, 37367–37384. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, M.; Zhao, Q.; Li, S.; Peng, Y.; Zhang, P.; Han, M. Vascular endothelial growth factor plays a critical role in the formation of the pre-metastatic niche via prostaglandin E2. Oncol. Rep. 2014, 32, 2477–2484. [Google Scholar] [CrossRef]
- Accioly, M.T.; Pacheco, P.; Maya-Monteiro, C.M.; Carrossini, N.; Robbs, B.K.; Oliveira, S.S.; Kaufmann, C.; Morgado-Diaz, J.A.; Bozza, P.T.; Viola, J.P.B. Lipid Bodies Are Reservoirs of Cyclooxygenase-2 and Sites of Prostaglandin-E2 Synthesis in Colon Cancer Cells. Cancer Res. 2008, 68, 1732–1740. [Google Scholar] [CrossRef]
- Shang, C.; Qiao, J.; Guo, H. The dynamic behavior of lipid droplets in the pre-metastatic niche. Cell Death Dis. 2020, 11, 990. [Google Scholar] [CrossRef]
- Li, P.; Lu, M.; Shi, J.; Gong, Z.; Hua, L.; Li, Q.; Lim, B.; Zhang, X.H.; Chen, X.; Li, S.; et al. Lung mesenchymal cells elicit lipid storage in neutrophils that fuel breast cancer lung metastasis. Nat. Immunol. 2020, 21, 1444–1455. [Google Scholar] [CrossRef]
- Diem, S.; Schmid, S.; Krapf, M.; Flatz, L.; Born, D.; Jochum, W.; Templeton, A.J.; Früh, M. Neutrophil-to-Lymphocyte ratio (NLR) and Platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lunge Cancer 2017, 111, 176–181. [Google Scholar] [CrossRef]
- Aloe, C.; Wang, H.; Vlahos, R.; Irving, L.; Steinfort, D.; Bozinovski, S. Emerging and multifaceted role of neutrophils in lung cancer. Transl. Lunge Cancer Res. 2021, 10, 2806–2818. [Google Scholar] [CrossRef] [PubMed]
- Eruslanov, E.B.; Bhojnagarwala, P.S.; Quatromoni, J.G.; Stephen, T.L.; Ranganathan, A.; Deshpande, C.; Akimova, T.; Vachani, A.; Litzky, L.; Hancock, W.W.; et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Investig. 2014, 124, 5466–5480. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Mishalian, I.; Bayuh, R.; Levy, L.; Zolotarov, L.; Michaeli, J.; Fridlender, Z.G. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunol. Immunother 2013, 62, 1745–1756. [Google Scholar] [CrossRef]
- Florea, A.-M.; Büsselberg, D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancer 2011, 3, 1351–1371. [Google Scholar] [CrossRef]
- D’Arpa, P.; Liu, L.F. Topoisomerase-targeting antitumor drugs. Biochim. Biophys. Acta (BBA)-Rev. Cancer 1989, 989, 163–177. [Google Scholar] [CrossRef]
- Janciauskiene, S.; Tumpara, S.; Wiese, M.; Wrenger, S.; Vijayan, V.; Gueler, F.; Chen, R.; Madyaningrana, K.; Mahadeva, R.; Welte, T.; et al. Alpha1-antitrypsin binds hemin and prevents oxidative activation of human neutrophils: Putative pathophysiological significance. J. Leukoc. Biol. 2017, 102, 1127–1141. [Google Scholar] [CrossRef]
- Ercetin, E.; Richtmann, S.; Delgado, B.M.; Gomez-Mariano, G.; Wrenger, S.; Korenbaum, E.; Liu, B.; DeLuca, D.; Kühnel, M.P.; Jonigk, D.; et al. Clinical Significance of SERPINA1 Gene and Its Encoded Alpha1-antitrypsin Protein in NSCLC. Cancer 2019, 11, 1306. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wajid, J. EnrichR: Provides an R Interface to All ‘Enrichr’ Databases. Available online: https://CRAN.R-project.org/package=enrichR (accessed on 15 April 2022).
- Wickham, H. Ggplot2. Wiley interdisciplinary reviews: Computational statistics. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Slowikowski, K.; Hughes, S.; Lukauskas, S.; Irisson, J.-O.; Kamvar, Z.N.; Ryan, T.; Christopher, D.; Hiroaki, Y.; Gramme, P. Package Ggrepel. Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. 2018. Available online: https://CRAN.R-project.org/package=ggrepel (accessed on 18 March 2021).
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R. package 2015, 1, 790. [Google Scholar]
- Schneider, M.A.; Granzow, M.; Warth, A.; Schnabel, P.A.; Thomas, M.; Herth, F.J.F.; Dienemann, H.; Muley, T.; Meister, M. Glycodelin: A New Biomarker with Immunomodulatory Functions in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 3529–3540. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumor marker prognostic studies (REMARK). Breast Cancer Res. Treat 2006, 100, 229–235. [Google Scholar] [CrossRef]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Győrffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, 51862. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Kettunen, M.I.; Hu, D.-E.; Brindle, K.M. 1H MRS-visible lipids accumulate during apoptosis of lymphoma cells in vitro and in vivo. Magn. Reason. Med. 2005, 54, 43–50. [Google Scholar] [CrossRef]
- Sun, Y.; He, W.; Luo, M.; Zhou, Y.; Chang, G.; Ren, W.; Wu, K.; Li, X.; Shen, J.; Zhao, X.; et al. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 2015, 36, 4133–4141. [Google Scholar] [CrossRef]
- Gao, Y.; Nan, X.; Shi, X.; Mu, X.; Liu, B.; Zhu, H.; Yao, B.; Liu, X.; Yang, T.; Hu, Y. SREBP1 promotes the invasion of colorectal cancer accompanied upregulation of MMP7 expression and NF-κB pathway activation. BMC Cancer 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef]
- Wu, L.; Saxena, S.; Singh, R.K. Neutrophils in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1224, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, C.C.; Malanchi, I. Neutrophils in cancer: Heterogeneous and multifaceted. Nat. Rev. Immunol. 2022, 22, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Carus, A.; Ladekarl, M.; Hager, H.; Nedergaard, B.S.; Donskov, F. Tumour-associated CD66b+ neutrophil count is an independent prognostic factor for recurrence in localised cervical cancer. Br. J. Cancer 2013, 108, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- Rakaee, M.; Busund, L.T.; Paulsen, E.E.; Richardsen, E.; Al-Saad, S.; Andersen, S.; Donnem, T.; Bremnes, R.M.; Kilvaer, T.K. Prognostic effect of intratumoral neutrophils across histological subtypes of non-small cell lung cancer. Oncotarget 2016, 7, 72184–72196. [Google Scholar] [CrossRef]
- D’Avila, H.; Roque, N.R.; Cardoso, R.M.; Castro-Faria-Neto, H.C.; Melo, R.C.N.; Bozza, P.T. Neutrophils recruited to the site of Mycobacterium bovis BCG infection undergo apoptosis and modulate lipid body biogenesis and prostaglandin E2 production by macrophages. Cell Microbiol. 2008, 10, 2589–2604. [Google Scholar] [CrossRef]
- Jin, C.; Yuan, P. Implications of lipid droplets in lung cancer: Associations with drug resistance. Oncol. Lett. 2020, 20, 2091–2104. [Google Scholar] [CrossRef]
- Herms, A.; Bosch, M.; Reddy, B.J.; Schieber, N.L.; Fajardo, A.; Rupérez, C.; Fernández-Vidal, A.; Ferguson, C.; Rentero, C.; Tebar, F.; et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 2015, 6, 7176. [Google Scholar] [CrossRef]
- Eichelberger, K.R.; Goldman, W.E. Manipulating neutrophil degranulation as a bacterial virulence strategy. PLoS Pathog. 2020, 16, e1009054. [Google Scholar] [CrossRef]
- Petan, T.; Jarc, E.; Jusović, M. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Corazao-Rozas, P.; Guerreschi, P.; André, F.; Gabert, P.E.; Lancel, S.; Dekiouk, S.; Fontaine, D.; Tardivel, M.; Savina, A.; Quesnel, B.; et al. Mitochondrial oxidative phosphorylation controls cancer cell’s life and death decisions upon exposure to MAPK inhibitors. Oncotarget 2016, 7, 39473–39485. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. Febs. J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Cheng, X.; Wu, X.; Yoo, J.Y.; Cheng, C.; Guo, J.Y.; Mo, X.; Ru, P.; Hurwitz, B.; Kim, S.-H.; et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1–Mediated Lipogenesis. Clin. Cancer Res. 2016, 22, 5337–5348. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Cruz, A.; Barbosa, J.; Bonifácio, V.D.B.; Pinto, S.N. Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics. Molecules 2022, 27, 991. [Google Scholar] [CrossRef] [PubMed]
- Alpsoy, A.; Yasa, S.; Gündüz, U. Etoposide resistance in MCF-7 breast cancer cell line is marked by multiple mechanisms. Biomed. Pharmacother 2014, 68, 351–355. [Google Scholar] [CrossRef]
- Liu, K.; Graves, J.D.; Lin, F.-T.; Lin, W.-C. Overexpression of TopBP1, a canonical ATR/Chk1 activator, paradoxically hinders ATR/Chk1 activation in cancer. J. Biol. Chem. 2021, 296, 100382. [Google Scholar] [CrossRef]
- Georgakilas, A.G.; Martin, O.A.; Bonner, W.M. P21: A Two-Faced Genome Guardian. Trends Mol. Med. 2017, 23, 310–319. [Google Scholar] [CrossRef]
- Triantafyllou, E.A.; Georgatsou, E.; Mylonis, I.; Simos, G.; Paraskeva, E. Expression of AGPAT2, an enzyme involved in the glycerophospholipid/triacylglycerol biosynthesis pathway, is directly regulated by HIF-1 and promotes survival and etoposide resistance of cancer cells under hypoxia. Biochim. Biophys. Acta Mol. Cell Biol. Lipid 2018, 1863, 1142–1152. [Google Scholar] [CrossRef]
- Zamagni, A.; Pasini, A.; Pirini, F.; Ravaioli, S.; Giordano, E.; Tesei, A.; Calistri, D.; Ulivi, P.; Fabbri, F.; Foca, F.; et al. CDKN1A upregulation and cisplatin-pemetrexed resistance in non-small cell lung cancer cells. Int. J. Oncol. 2020, 56, 1574–1584. [Google Scholar] [CrossRef]
- Kong, H.J.; Kwon, E.J.; Kwon, O.S.; Lee, H.; Choi, J.Y.; Kim, Y.J.; Kim, W.; Cha, H.J. Crosstalk between YAP and TGFβ regulates SERPINE1 expression in mesenchymal lung cancer cells. Int. J. Oncol. 2021, 58, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Pavón, M.A.; Arroyo-Solera, I.; Téllez-Gabriel, M.; León, X.; Virós, D.; López, M.; Gallardo, A.; Céspedes, M.V.; Casanova, I.; López-Pousa, A.; et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015, 6, 29016–29033. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Virshup, D.M. Wnt Signaling and Drug Resistance in Cancer. Mol. Pharmacol. 2020, 97, 72–89. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Bronson, S.M.; Pal-Nath, D.; Miller, T.W.; Soto-Pantoja, D.R.; Roberts, D.D. Functions of Thrombospondin-1 in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 4570. [Google Scholar] [CrossRef]
- Ruiz, C.F.; Montal, E.D.; Haley, J.A.; Haley, J.D. SREBP1 regulates mitochondrial metabolism in oncogenic KRAS expressing NSCLC. FASEB J. 2020, 34, 10574–10589. [Google Scholar] [CrossRef]
- Li, L.Y.; Yang, Q.; Jiang, Y.Y.; Yang, W.; Jiang, Y.; Li, X.; Hazawa, M.; Zhou, B.; Huang, G.W.; Xu, X.E.; et al. Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat. Commun. 2021, 12, 4362. [Google Scholar] [CrossRef] [PubMed]
- Im, S.S.; Yousef, L.; Blaschitz, C.; Liu, J.Z.; Edwards, R.A.; Young, S.G.; Raffatellu, M.; Osborne, T.F. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011, 13, 540–549. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Chen, L.; Ma, W.L.; Cheng, W.C.; Yang, J.C.; Wang, H.C.; Su, Y.T.; Ahmad, A.; Hung, Y.C.; Chang, W.C. Targeting lipid droplet lysophosphatidylcholine for cisplatin chemotherapy. J. Cell Mol. Med. 2020, 24, 7187–7200. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Luo, X. Lipid droplet and its implication in cancer progression. Am. J. Cancer Res. 2020, 10, 4112–4122. [Google Scholar]








| Gene Name | Base Mean | Log2 Fold Change | LfcSE | Stat | p-Value | Padj | Direction |
|---|---|---|---|---|---|---|---|
| FGF1 | 36.33 | −6.67 | 1.08 | −6.17 | 6.71 × 10−10 | 1.68 × 10−10 | ▼ |
| ABCG1 | 937.21 | 6.65 | 0.22 | 30.56 | 4.10 × 10−205 | 4.10 × 10−204 | ▲ |
| MVD | 4164.03 | −3.51 | 0.14 | −25.53 | 9.40 × 10−144 | 4.50 × 10−143 | ▼ |
| APOE | 368.96 | −3.23 | 0.24 | −13.69 | 1.16 × 10−42 | 9.48 × 10−43 | ▼ |
| FASN | 26,278.63 | −3.08 | 0.15 | −20.98 | 9.99 × 10−98 | 2.49 × 10−97 | ▼ |
| SREBF1 | 6809.99 | −2.99 | 0.11 | −26.62 | 4.10 × 10−156 | 2.20 × 10−155 | ▼ |
| FDPS | 14,122.46 | −2.94 | 0.09 | −32.41 | 2.20 × 10−230 | 2.70 × 10−229 | ▼ |
| SCD | 105,179.20 | −2.90 | 0.13 | −22.52 | 2.40 × 10−112 | 7.50 × 10−112 | ▼ |
| LSS | 7094.47 | −2.67 | 0.12 | −22.58 | 6.60 × 10−113 | 2.10 × 10−112 | ▼ |
| FDFT1 | 9258.84 | −2.44 | 0.10 | −25.10 | 4.60 × 10−139 | 2.10 × 10−138 | ▼ |
| MVK | 836.61 | −1.92 | 0.12 | −15.87 | 1.02 × 10−56 | 1.19 × 10−56 | ▼ |
| IDI1 | 7365.73 | −1.74 | 0.12 | −14.88 | 4.35 × 10−50 | 4.31 × 10−50 | ▼ |
| HMGCS1 | 11,484.75 | −1.63 | 0.11 | −15.14 | 8.61 × 10−52 | 8.88 × 10−52 | ▼ |
| TM7SF2 | 754.88 | −1.53 | 0.17 | −9.05 | 1.48 × 10−19 | 5.72 × 10−20 | ▼ |
| NFYB | 1985.44 | 1.38 | 0.11 | 13.10 | 3.26 × 10−39 | 2.44 × 10−39 | ▲ |
| HMGCR | 8898.55 | −1.26 | 0.08 | −15.10 | 1.57 × 10−51 | 1.60 × 10−51 | ▼ |
| DHCR7 | 3593.68 | −1.16 | 0.10 | −11.66 | 2.03 × 10−31 | 1.20 × 10−31 | ▼ |
| ELOVL6 | 3212.98 | 1.15 | 0.10 | 11.23 | 2.85 × 10−29 | 1.58 × 10−29 | ▲ |
| RAN | 11,606.11 | −1.08 | 0.12 | −9.29 | 1.49 × 10−20 | 6.01 × 10−21 | ▼ |
| ERLIN2 | 2056.03 | 0.96 | 0.10 | 9.35 | 8.39 × 10−21 | 3.43 × 10−21 | ▲ |
| MBTPS2 | 2285.09 | 0.92 | 0.10 | 9.14 | 6.18 × 10−20 | 2.43 × 10−20 | ▲ |
| CYP51A1 | 5903.00 | −0.92 | 0.09 | −9.68 | 3.80 × 10−22 | 1.64 × 10−22 | ▼ |
| SEC14L2 | 773.96 | 0.81 | 0.11 | 7.62 | 2.44 × 10−14 | 7.60 × 10−15 | ▲ |
| SQLE | 7182.00 | −0.79 | 0.10 | −7.92 | 2.30 × 10−15 | 7.49 × 10−16 | ▼ |
| GPAM | 580.32 | −0.77 | 0.13 | −5.95 | 2.67 × 10−9 | 6.48 × 10−10 | ▼ |
| SREBF2 | 6029.96 | −0.65 | 0.09 | −7.29 | 3.10 × 10−13 | 9.17 × 10−14 | ▼ |
| SCAP | 2424.48 | 0.62 | 0.11 | 5.81 | 6.10 × 10−9 | 1.46 × 10−9 | ▲ |
| SC5D | 3318.08 | 0.60 | 0.10 | 6.27 | 3.73 × 10−10 | 9.45 × 10−11 | ▲ |
| PRKAA1 | 4033.46 | −0.59 | 0.11 | −5.30 | 1.16 × 10−7 | 2.60 × 10−8 | ▼ |
| KPNB1 | 26,881.55 | −0.57 | 0.09 | −6.04 | 1.51 × 10−9 | 3.73 × 10−10 | ▼ |
| SP1 | 5089.01 | −0.56 | 0.09 | −6.35 | 2.13 × 10−10 | 5.48 × 10−11 | ▼ |
| GGPS1 | 1151.02 | 0.54 | 0.11 | 4.75 | 2.07 × 10−6 | 4.35 × 10−7 | ▲ |
| ERLIN1 | 1600.73 | −0.51 | 0.12 | −4.32 | 1.59 × 10−5 | 3.17 × 10−6 | ▼ |
| NFYA | 1415.55 | −0.41 | 0.11 | −3.77 | 1.66 × 10−4 | 3.10 × 10−5 | ▼ |
| ACACA | 5645.03 | 0.41 | 0.12 | 3.49 | 4.78 × 10−4 | 8.65 × 10−5 | ▲ |
| SOD1 | 8249.72 | 0.37 | 0.12 | 3.14 | 1.71 × 10−3 | 2.97 × 10−4 | ▲ |
| NFYC | 2042.66 | −0.29 | 0.09 | −3.12 | 1.82 × 10−3 | 3.16 × 10−4 | ▼ |
| MBTPS1 | 6905.10 | −0.25 | 0.08 | −3.04 | 2.37 × 10−3 | 4.07 × 10−4 | ▼ |
| ACACB | 578.19 | 0.23 | 0.13 | 1.80 | 7.18 × 10−2 | 1.07 × 10−2 | ▲ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gründing, A.R.; Schneider, M.A.; Richtmann, S.; Kriegsmann, M.; Winter, H.; Martinez-Delgado, B.; Varona, S.; Liu, B.; DeLuca, D.S.; Held, J.; et al. Lung Adenocarcinoma Cell Sensitivity to Chemotherapies: A Spotlight on Lipid Droplets and SREBF1 Gene. Cancers 2022, 14, 4454. https://doi.org/10.3390/cancers14184454
Gründing AR, Schneider MA, Richtmann S, Kriegsmann M, Winter H, Martinez-Delgado B, Varona S, Liu B, DeLuca DS, Held J, et al. Lung Adenocarcinoma Cell Sensitivity to Chemotherapies: A Spotlight on Lipid Droplets and SREBF1 Gene. Cancers. 2022; 14(18):4454. https://doi.org/10.3390/cancers14184454
Chicago/Turabian StyleGründing, Anna Ricarda, Marc A. Schneider, Sarah Richtmann, Mark Kriegsmann, Hauke Winter, Beatriz Martinez-Delgado, Sarai Varona, Bin Liu, David S. DeLuca, Julia Held, and et al. 2022. "Lung Adenocarcinoma Cell Sensitivity to Chemotherapies: A Spotlight on Lipid Droplets and SREBF1 Gene" Cancers 14, no. 18: 4454. https://doi.org/10.3390/cancers14184454
APA StyleGründing, A. R., Schneider, M. A., Richtmann, S., Kriegsmann, M., Winter, H., Martinez-Delgado, B., Varona, S., Liu, B., DeLuca, D. S., Held, J., Wrenger, S., Muley, T., Meister, M., Welte, T., & Janciauskiene, S. (2022). Lung Adenocarcinoma Cell Sensitivity to Chemotherapies: A Spotlight on Lipid Droplets and SREBF1 Gene. Cancers, 14(18), 4454. https://doi.org/10.3390/cancers14184454